METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis
- PMID: 35012593
- PMCID: PMC8744223
- DOI: 10.1186/s13046-021-02227-8
METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis
Abstract
Background: Colorectal carcinoma (CRC) is the third most common cancer and second most common cause of cancer-related deaths worldwide. Ribonucleic acid (RNA) N6-methyladnosine (m6A) and methyltransferase-like 3 (METTL3) play key roles in cancer progression. However, the roles of m6A and METTL3 in CRC progression require further clarification.
Methods: Adenoma and CRC samples were examined to detect m6A and METTL3 levels, and tissue microarrays were performed to evaluate the association of m6A and METTL3 levels with the survival of patients with CRC. The biological functions of METTL3 were investigated through cell counting kit-8, wound healing, and transwell assays. M6A epitranscriptomic microarray, methylated RNA immunoprecipitation-qPCR, RNA stability, luciferase reporter, and RNA immunoprecipitation assays were performed to explore the mechanism of METTL3 in CRC progression.
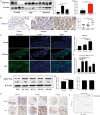
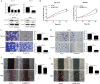
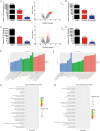
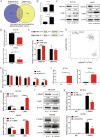
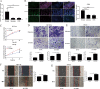
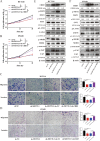
Results: M6A and METTL3 levels were substantially elevated in CRC tissues, and patients with CRC with a high m6A or METTL3 levels exhibited shorter overall survival. METTL3 knockdown substantially inhibited the proliferation, migration, and invasion of CRC cells. An m6A epitranscriptomic microarray revealed that the cell polarity regulator Crumbs3 (CRB3) was the downstream target of METTL3. METTL3 knockdown substantially reduced the m6A level of CRB3, and inhibited the degradation of CRB3 mRNA to increase CRB3 expression. Luciferase reporter assays also showed that the transcriptional level of wild-type CRB3 significantly increased after METTL3 knockdown but not its level of variation. Knockdown of YT521-B homology domain-containing family protein 2 (YTHDF2) substantially increased CRB3 expression. RNA immunoprecipitation assays also verified the direct interaction between the YTHDF2 and CRB3 mRNA, and this direct interaction was impaired after METTL3 inhibition. In addition, CRB3 knockdown significantly promoted the proliferation, migration, and invasion of CRC cells. Mechanistically, METTL3 knockdown activated the Hippo pathway and reduced nuclear localization of Yes1-associated transcriptional regulator, and the effects were reversed by CRB3 knockdown.
Conclusions: M6A and METTL3 levels were substantially elevated in CRC tissues relative to normal tissues. Patients with CRC with high m6A or METTL3 levels exhibited shorter overall survival, and METTL3 promoted CRC progression. Mechanistically, METTL3 regulated the progression of CRC by regulating the m6A-CRB3-Hippo pathway.
Keywords: Colorectal carcinoma; Crumbs3; Hippo pathway; Methyltransferase-like 3; RNA N6-methyladnosine.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

