Transcriptomic analysis of Pseudomonas ogarae F113 reveals the antagonistic roles of AmrZ and FleQ during rhizosphere adaption
- PMID: 35012704
- PMCID: PMC8914362
- DOI: 10.1099/mgen.0.000750
Transcriptomic analysis of Pseudomonas ogarae F113 reveals the antagonistic roles of AmrZ and FleQ during rhizosphere adaption
Abstract
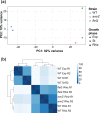
Rhizosphere colonization by bacteria involves molecular and cellular mechanisms, such as motility and chemotaxis, biofilm formation, metabolic versatility, or biosynthesis of secondary metabolites, among others. Nonetheless, there is limited knowledge concerning the main regulatory factors that drive the rhizosphere colonization process. Here we show the importance of the AmrZ and FleQ transcription factors for adaption in the plant growth-promoting rhizobacterium (PGPR) and rhizosphere colonization model Pseudomonas ogarae F113. RNA-Seq analyses of P. ogarae F113 grown in liquid cultures either in exponential and stationary growth phase, and rhizosphere conditions, revealed that rhizosphere is a key driver of global changes in gene expression in this bacterium. Regarding the genetic background, this work has revealed that a mutation in fleQ causes considerably more alterations in the gene expression profile of this bacterium than a mutation in amrZ under rhizosphere conditions. The functional analysis has revealed that in P. ogarae F113, the transcription factors AmrZ and FleQ regulate genes involved in diverse bacterial functions. Notably, in the rhizosphere, these transcription factors antagonistically regulate genes related to motility, biofilm formation, nitrogen, sulfur, and amino acid metabolism, transport, signalling, and secretion, especially the type VI secretion systems. These results define the regulon of two important bifunctional transcriptional regulators in pseudomonads during the process of rhizosphere colonization.
Keywords: AmrZ; FleQ; Pseudomonas ogarae; regulation; rhizosphere; transcriptomics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

