Diagnosis of Genetic White Matter Disorders by Singleton Whole-Exome and Genome Sequencing Using Interactome-Driven Prioritization
- PMID: 35012964
- PMCID: PMC8901178
- DOI: 10.1212/WNL.0000000000013278
Diagnosis of Genetic White Matter Disorders by Singleton Whole-Exome and Genome Sequencing Using Interactome-Driven Prioritization
Abstract
Background and objectives: Genetic white matter disorders (GWMD) are of heterogeneous origin, with >100 causal genes identified to date. Classic targeted approaches achieve a molecular diagnosis in only half of all patients. We aimed to determine the clinical utility of singleton whole-exome sequencing and whole-genome sequencing (sWES-WGS) interpreted with a phenotype- and interactome-driven prioritization algorithm to diagnose GWMD while identifying novel phenotypes and candidate genes.
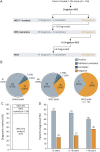
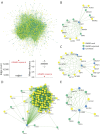
Methods: A case series of patients of all ages with undiagnosed GWMD despite extensive standard-of-care paraclinical studies were recruited between April 2017 and December 2019 in a collaborative study at the Bellvitge Biomedical Research Institute (IDIBELL) and neurology units of tertiary Spanish hospitals. We ran sWES and WGS and applied our interactome-prioritization algorithm based on the network expansion of a seed group of GWMD-related genes derived from the Human Phenotype Ontology terms of each patient.
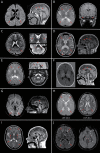
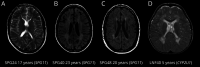
Results: We evaluated 126 patients (101 children and 25 adults) with ages ranging from 1 month to 74 years. We obtained a first molecular diagnosis by singleton WES in 59% of cases, which increased to 68% after annual reanalysis, and reached 72% after WGS was performed in 16 of the remaining negative cases. We identified variants in 57 different genes among 91 diagnosed cases, with the most frequent being RNASEH2B, EIF2B5, POLR3A, and PLP1, and a dual diagnosis underlying complex phenotypes in 6 families, underscoring the importance of genomic analysis to solve these cases. We discovered 9 candidate genes causing novel diseases and propose additional putative novel candidate genes for yet-to-be discovered GWMD.
Discussion: Our strategy enables a high diagnostic yield and is a good alternative to trio WES/WGS for GWMD. It shortens the time to diagnosis compared to the classical targeted approach, thus optimizing appropriate management. Furthermore, the interactome-driven prioritization pipeline enables the discovery of novel disease-causing genes and phenotypes, and predicts novel putative candidate genes, shedding light on etiopathogenic mechanisms that are pivotal for myelin generation and maintenance.
Copyright © 2022 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Figures