The transcription factor ZIP-1 promotes resistance to intracellular infection in Caenorhabditis elegans
- PMID: 35013162
- PMCID: PMC8748929
- DOI: 10.1038/s41467-021-27621-w
The transcription factor ZIP-1 promotes resistance to intracellular infection in Caenorhabditis elegans
Abstract
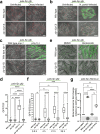
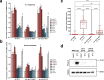
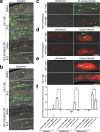
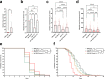
Defense against intracellular infection has been extensively studied in vertebrate hosts, but less is known about invertebrate hosts; specifically, the transcription factors that induce defense against intracellular intestinal infection in the model nematode Caenorhabditis elegans remain understudied. Two different types of intracellular pathogens that naturally infect the C. elegans intestine are the Orsay virus, which is an RNA virus, and microsporidia, which comprise a phylum of fungal pathogens. Despite their molecular differences, these pathogens induce a common host transcriptional response called the intracellular pathogen response (IPR). Here we show that zip-1 is an IPR regulator that functions downstream of all known IPR-activating and regulatory pathways. zip-1 encodes a putative bZIP transcription factor, and we show that zip-1 controls induction of a subset of genes upon IPR activation. ZIP-1 protein is expressed in the nuclei of intestinal cells, and is at least partially required in the intestine to upregulate IPR gene expression. Importantly, zip-1 promotes resistance to infection by the Orsay virus and by microsporidia in intestinal cells. Altogether, our results indicate that zip-1 represents a central hub for triggers of the IPR, and that this transcription factor has a protective function against intracellular pathogen infection in C. elegans.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Yamada, T. et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 10.1038/s41590-021-00942-0 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

