An assembly of nuclear bodies associates with the active VSG expression site in African trypanosomes
- PMID: 35013170
- PMCID: PMC8748868
- DOI: 10.1038/s41467-021-27625-6
An assembly of nuclear bodies associates with the active VSG expression site in African trypanosomes
Abstract
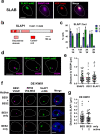
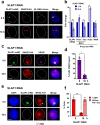
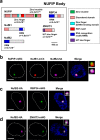
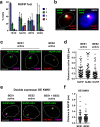
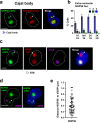
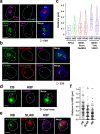
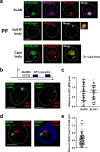
A Variant Surface Glycoprotein (VSG) coat protects bloodstream form Trypanosoma brucei. Prodigious amounts of VSG mRNA (~7-10% total) are generated from a single RNA polymerase I (Pol I) transcribed VSG expression site (ES), necessitating extremely high levels of localised splicing. We show that splicing is required for processive ES transcription, and describe novel ES-associated T. brucei nuclear bodies. In bloodstream form trypanosomes, the expression site body (ESB), spliced leader array body (SLAB), NUFIP body and Cajal bodies all frequently associate with the active ES. This assembly of nuclear bodies appears to facilitate the extraordinarily high levels of transcription and splicing at the active ES. In procyclic form trypanosomes, the NUFIP body and SLAB do not appear to interact with the Pol I transcribed procyclin locus. The congregation of a restricted number of nuclear bodies at a single active ES, provides an attractive mechanism for how monoallelic ES transcription is mediated.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

