TASOR epigenetic repressor cooperates with a CNOT1 RNA degradation pathway to repress HIV
- PMID: 35013187
- PMCID: PMC8748822
- DOI: 10.1038/s41467-021-27650-5
TASOR epigenetic repressor cooperates with a CNOT1 RNA degradation pathway to repress HIV
Abstract
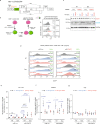
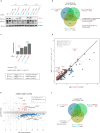
The Human Silencing Hub (HUSH) complex constituted of TASOR, MPP8 and Periphilin recruits the histone methyl-transferase SETDB1 to spread H3K9me3 repressive marks across genes and transgenes in an integration site-dependent manner. The deposition of these repressive marks leads to heterochromatin formation and inhibits gene expression, but the underlying mechanism is not fully understood. Here, we show that TASOR silencing or HIV-2 Vpx expression, which induces TASOR degradation, increases the accumulation of transcripts derived from the HIV-1 LTR promoter at a post-transcriptional level. Furthermore, using a yeast 2-hybrid screen, we identify new TASOR partners involved in RNA metabolism including the RNA deadenylase CCR4-NOT complex scaffold CNOT1. TASOR and CNOT1 synergistically repress HIV expression from its LTR. Similar to the RNA-induced transcriptional silencing complex found in fission yeast, we show that TASOR interacts with the RNA exosome and RNA Polymerase II, predominantly under its elongating state. Finally, we show that TASOR facilitates the association of RNA degradation proteins with RNA polymerase II and is detected at transcriptional centers. Altogether, we propose that HUSH operates at the transcriptional and post-transcriptional levels to repress HIV proviral expression.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Chougui G, et al. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018;3:891–897. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

