Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts
- PMID: 35013199
- PMCID: PMC8748880
- DOI: 10.1038/s41467-021-27674-x
Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts
Abstract
Cross-reactive immune responses to SARS-CoV-2 have been observed in pre-pandemic cohorts and proposed to contribute to host protection. Here we assess 52 COVID-19 household contacts to capture immune responses at the earliest timepoints after SARS-CoV-2 exposure. Using a dual cytokine FLISpot assay on peripheral blood mononuclear cells, we enumerate the frequency of T cells specific for spike, nucleocapsid, membrane, envelope and ORF1 SARS-CoV-2 epitopes that cross-react with human endemic coronaviruses. We observe higher frequencies of cross-reactive (p = 0.0139), and nucleocapsid-specific (p = 0.0355) IL-2-secreting memory T cells in contacts who remained PCR-negative despite exposure (n = 26), when compared with those who convert to PCR-positive (n = 26); no significant difference in the frequency of responses to spike is observed, hinting at a limited protective function of spike-cross-reactive T cells. Our results are thus consistent with pre-existing non-spike cross-reactive memory T cells protecting SARS-CoV-2-naïve contacts from infection, thereby supporting the inclusion of non-spike antigens in second-generation vaccines.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

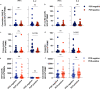
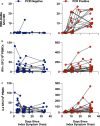
Comment in
-
Understanding Kids and COVID.Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2203753119. doi: 10.1073/pnas.2203753119. Epub 2022 Mar 22. Proc Natl Acad Sci U S A. 2022. PMID: 35316141 Free PMC article. No abstract available.
References
-
- Nelde, A. et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 10.1038/s41590-020-00808-x. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

