Functional dissection of inherited non-coding variation influencing multiple myeloma risk
- PMID: 35013207
- PMCID: PMC8748989
- DOI: 10.1038/s41467-021-27666-x
Functional dissection of inherited non-coding variation influencing multiple myeloma risk
Erratum in
-
Author Correction: Functional dissection of inherited non-coding variation influencing multiple myeloma risk.Nat Commun. 2022 Dec 13;13(1):7725. doi: 10.1038/s41467-022-35411-1. Nat Commun. 2022. PMID: 36513657 Free PMC article. No abstract available.
Abstract
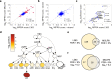
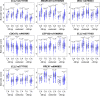
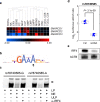
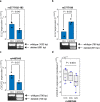
Thousands of non-coding variants have been associated with increased risk of human diseases, yet the causal variants and their mechanisms-of-action remain obscure. In an integrative study combining massively parallel reporter assays (MPRA), expression analyses (eQTL, meQTL, PCHiC) and chromatin accessibility analyses in primary cells (caQTL), we investigate 1,039 variants associated with multiple myeloma (MM). We demonstrate that MM susceptibility is mediated by gene-regulatory changes in plasma cells and B-cells, and identify putative causal variants at six risk loci (SMARCD3, WAC, ELL2, CDCA7L, CEP120, and PREX1). Notably, three of these variants co-localize with significant plasma cell caQTLs, signaling the presence of causal activity at these precise genomic positions in an endogenous chromosomal context in vivo. Our results provide a systematic functional dissection of risk loci for a hematologic malignancy.
© 2022. The Author(s).
Conflict of interest statement
Authors T.O., O.M., G.H.H., G.T., G.L.N., K.G., I.J., T.R., U.T., and K.S. are employed by deCODE Genetics/Amgen Inc. The remaining authors declare no competing interests.
Figures




References
-
- Pertesi, M. et al. Genetic predisposition for multiple myeloma. Leukemia10.1038/s41375-019-0703-6 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

