Age-dependent pathogenic characteristics of SARS-CoV-2 infection in ferrets
- PMID: 35013229
- PMCID: PMC8748994
- DOI: 10.1038/s41467-021-27717-3
Age-dependent pathogenic characteristics of SARS-CoV-2 infection in ferrets
Abstract
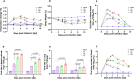
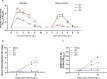
While the seroprevalence of SARS-CoV-2 in healthy people does not differ significantly among age groups, those aged 65 years or older exhibit strikingly higher COVID-19 mortality compared to younger individuals. To further understand differing COVID-19 manifestations in patients of different ages, three age groups of ferrets are infected with SARS-CoV-2. Although SARS-CoV-2 is isolated from all ferrets regardless of age, aged ferrets (≥3 years old) show higher viral loads, longer nasal virus shedding, and more severe lung inflammatory cell infiltration, and clinical symptoms compared to juvenile (≤6 months) and young adult (1-2 years) groups. Furthermore, direct contact ferrets co-housed with the virus-infected aged group shed more virus than direct-contact ferrets co-housed with virus-infected juvenile or young adult ferrets. Transcriptome analysis of aged ferret lungs reveals strong enrichment of gene sets related to type I interferon, activated T cells, and M1 macrophage responses, mimicking the gene expression profile of severe COVID-19 patients. Thus, SARS-CoV-2-infected aged ferrets highly recapitulate COVID-19 patients with severe symptoms and are useful for understanding age-associated infection, transmission, and pathogenesis of SARS-CoV-2.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Age-dependent pathogenic characteristics of SARS-CoV-2 infection in ferrets.Res Sq [Preprint]. 2021 Mar 29:rs.3.rs-131380. doi: 10.21203/rs.3.rs-131380/v2. Res Sq. 2021. Update in: Nat Commun. 2022 Jan 10;13(1):21. doi: 10.1038/s41467-021-27717-3. PMID: 33821260 Free PMC article. Updated. Preprint.
Similar articles
-
Age-dependent pathogenic characteristics of SARS-CoV-2 infection in ferrets.Res Sq [Preprint]. 2021 Mar 29:rs.3.rs-131380. doi: 10.21203/rs.3.rs-131380/v2. Res Sq. 2021. Update in: Nat Commun. 2022 Jan 10;13(1):21. doi: 10.1038/s41467-021-27717-3. PMID: 33821260 Free PMC article. Updated. Preprint.
-
Age-Related Susceptibility of Ferrets to SARS-CoV-2 Infection.J Virol. 2022 Feb 9;96(3):e0145521. doi: 10.1128/JVI.01455-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34817200 Free PMC article.
-
Critical role of neutralizing antibody for SARS-CoV-2 reinfection and transmission.Emerg Microbes Infect. 2021 Dec;10(1):152-160. doi: 10.1080/22221751.2021.1872352. Emerg Microbes Infect. 2021. PMID: 33407005 Free PMC article.
-
Animal Models of COVID-19 II. Comparative Immunology.ILAR J. 2021 Dec 31;62(1-2):17-34. doi: 10.1093/ilar/ilab010. ILAR J. 2021. PMID: 33914873 Free PMC article. Review.
-
Experimental Models for SARS-CoV-2 Infection.Mol Cells. 2021 Jun 30;44(6):377-383. doi: 10.14348/molcells.2021.0094. Mol Cells. 2021. PMID: 34187969 Free PMC article. Review.
Cited by
-
Natural Transmission and Experimental Models of SARS CoV-2 Infection in Animals.Comp Med. 2021 Oct 1;71(5):369-382. doi: 10.30802/AALAS-CM-21-000046. Epub 2021 Oct 26. Comp Med. 2021. PMID: 34702427 Free PMC article.
-
SARS-CoV-2 variants with NSP12 P323L/G671S mutations display enhanced virus replication in ferret upper airways and higher transmissibility.Cell Rep. 2023 Sep 26;42(9):113077. doi: 10.1016/j.celrep.2023.113077. Epub 2023 Sep 6. Cell Rep. 2023. PMID: 37676771 Free PMC article.
-
Mechanism insights and therapeutic intervention of tumor metastasis: latest developments and perspectives.Signal Transduct Target Ther. 2024 Aug 2;9(1):192. doi: 10.1038/s41392-024-01885-2. Signal Transduct Target Ther. 2024. PMID: 39090094 Free PMC article. Review.
-
Recent review of COVID-19 management: diagnosis, treatment and vaccination.Pharmacol Rep. 2022 Dec;74(6):1120-1148. doi: 10.1007/s43440-022-00425-5. Epub 2022 Oct 10. Pharmacol Rep. 2022. PMID: 36214969 Free PMC article. Review.
-
The pathogenesis of coronavirus-19 disease.J Biomed Sci. 2022 Oct 26;29(1):87. doi: 10.1186/s12929-022-00872-5. J Biomed Sci. 2022. PMID: 36289507 Free PMC article. Review.
References
-
- Organization, W. H. WHO Director-General’s opening remarks at the media briefing on COVID-19-11 March 2020 (2020).
-
- Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. - PubMed
-
- ECDC. COVID-19 situation update worldwide, as of 25 November 2020. (2020).
Publication types
MeSH terms
Substances
Grants and funding
- 2020R1A2C3008339/National Research Foundation of Korea (NRF)
- KGM9942011/Korea Research Institute of Bioscience and Biotechnology (KRIBB)
- AI140705S/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 AI140705/AI/NIAID NIH HHS/United States
- R01 DE028521/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

