Proximity labeling identifies a repertoire of site-specific R-loop modulators
- PMID: 35013239
- PMCID: PMC8748879
- DOI: 10.1038/s41467-021-27722-6
Proximity labeling identifies a repertoire of site-specific R-loop modulators
Abstract
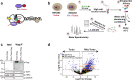
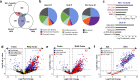
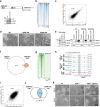
R-loops are three-stranded nucleic acid structures that accumulate on chromatin in neurological diseases and cancers and contribute to genome instability. Using a proximity-dependent labeling system, we identified distinct classes of proteins that regulate R-loops in vivo through different mechanisms. We show that ATRX suppresses R-loops by interacting with RNAs and preventing R-loop formation. Our proteomics screen also discovered an unexpected enrichment for proteins containing zinc fingers and homeodomains. One of the most consistently enriched proteins was activity-dependent neuroprotective protein (ADNP), which is frequently mutated in ASD and causal in ADNP syndrome. We find that ADNP resolves R-loops in vitro and that it is necessary to suppress R-loops in vivo at its genomic targets. Furthermore, deletion of the ADNP homeodomain severely diminishes R-loop resolution activity in vitro, results in R-loop accumulation at ADNP targets, and compromises neuronal differentiation. Notably, patient-derived human induced pluripotent stem cells that contain an ADNP syndrome-causing mutation exhibit R-loop and CTCF accumulation at ADNP targets. Our findings point to a specific role for ADNP-mediated R-loop resolution in physiological and pathological neuronal function and, more broadly, to a role for zinc finger and homeodomain proteins in R-loop regulation, with important implications for developmental disorders and cancers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Garcia-Muse T, Aguilera A. R loops: from physiological to pathological roles. Cell. 2019;179:604–618. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

