Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation
- PMID: 35013280
- PMCID: PMC8748714
- DOI: 10.1038/s41467-021-27760-0
Dynamics of GLP-1R peptide agonist engagement are correlated with kinetics of G protein activation
Abstract
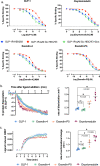
The glucagon-like peptide-1 receptor (GLP-1R) has broad physiological roles and is a validated target for treatment of metabolic disorders. Despite recent advances in GLP-1R structure elucidation, detailed mechanistic understanding of how different peptides generate profound differences in G protein-mediated signalling is still lacking. Here we combine cryo-electron microscopy, molecular dynamics simulations, receptor mutagenesis and pharmacological assays, to interrogate the mechanism and consequences of GLP-1R binding to four peptide agonists; glucagon-like peptide-1, oxyntomodulin, exendin-4 and exendin-P5. These data reveal that distinctions in peptide N-terminal interactions and dynamics with the GLP-1R transmembrane domain are reciprocally associated with differences in the allosteric coupling to G proteins. In particular, transient interactions with residues at the base of the binding cavity correlate with enhanced kinetics for G protein activation, providing a rationale for differences in G protein-mediated signalling efficacy from distinct agonists.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


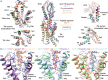





References
-
- Nauck MA, Meier JJ. MANAGEMENT OF ENDOCRINE DISEASE: are all GLP-1 agonists equal in the treatment of type 2 diabetes? Eur. J. Endocrinol. 2019;181:R211–R234. - PubMed
-
- Wootten D, et al. Differential activation and modulation of the glucagon-like peptide-1 receptor by small molecule ligands. Mol. Pharm. 2013;83:822–834. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

