Spatial transcriptomics using combinatorial fluorescence spectral and lifetime encoding, imaging and analysis
- PMID: 35013281
- PMCID: PMC8748653
- DOI: 10.1038/s41467-021-27798-0
Spatial transcriptomics using combinatorial fluorescence spectral and lifetime encoding, imaging and analysis
Abstract
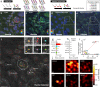
Multiplexed mRNA profiling in the spatial context provides new information enabling basic research and clinical applications. Unfortunately, existing spatial transcriptomics methods are limited due to either low multiplexing or complexity. Here, we introduce a spatialomics technology, termed Multi Omic Single-scan Assay with Integrated Combinatorial Analysis (MOSAICA), that integrates in situ labeling of mRNA and protein markers in cells or tissues with combinatorial fluorescence spectral and lifetime encoded probes, spectral and time-resolved fluorescence imaging, and machine learning-based decoding. We demonstrate MOSAICA's multiplexing scalability in detecting 10-plex targets in fixed colorectal cancer cells using combinatorial labeling of five fluorophores with facile error-detection and removal of autofluorescence. MOSAICA's analysis is strongly correlated with sequencing data (Pearson's r = 0.96) and was further benchmarked using RNAscopeTM and LGC StellarisTM. We further apply MOSAICA for multiplexed analysis of clinical melanoma Formalin-Fixed Paraffin-Embedded (FFPE) tissues. We finally demonstrate simultaneous co-detection of protein and mRNA in cancer cells.
© 2022. The Author(s).
Conflict of interest statement
T.V. and A.V. are now Arvetas Biosciences Inc employees. W.Z. is a cofounder of Velox Biosystems Inc., Amberstone Biosciences Inc., and Arvetas Biosciences Inc. P.N.H. holds a part-time position at Amberstone Biosciences Inc. A.G. is a founder of Alyra therapeutics. The remaining authors declare no competing interests.
Figures





References
-
- Pichon X, Lagha M, Mueller F, Bertrand E. A Growing Toolbox to Image Gene Expression in Single Cells: Sensitive Approaches for Demanding Challenges. Mol. Cell. 2018;71:468–480. - PubMed
-
- Barsoum I, Tawedrous E, Faragalla H, Yousef GM. Histo-genomics: digital pathology at the forefront of precision medicine. Diagnosis. 2018;6:203–212. - PubMed
-
- Gryglewski G, et al. Spatial analysis and high resolution mapping of the human whole-brain transcriptome for integrative analysis in neuroimaging. Neuroimage. 2018;176:259–267. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

