Hypocrates is a genetically encoded fluorescent biosensor for (pseudo)hypohalous acids and their derivatives
- PMID: 35013284
- PMCID: PMC8748444
- DOI: 10.1038/s41467-021-27796-2
Hypocrates is a genetically encoded fluorescent biosensor for (pseudo)hypohalous acids and their derivatives
Abstract
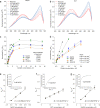
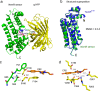
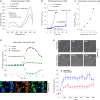
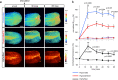
The lack of tools to monitor the dynamics of (pseudo)hypohalous acids in live cells and tissues hinders a better understanding of inflammatory processes. Here we present a fluorescent genetically encoded biosensor, Hypocrates, for the visualization of (pseudo)hypohalous acids and their derivatives. Hypocrates consists of a circularly permuted yellow fluorescent protein integrated into the structure of the transcription repressor NemR from Escherichia coli. We show that Hypocrates is ratiometric, reversible, and responds to its analytes in the 106 M-1s-1 range. Solving the Hypocrates X-ray structure provided insights into its sensing mechanism, allowing determination of the spatial organization in this circularly permuted fluorescent protein-based redox probe. We exemplify its applicability by imaging hypohalous stress in bacteria phagocytosed by primary neutrophils. Finally, we demonstrate that Hypocrates can be utilized in combination with HyPerRed for the simultaneous visualization of (pseudo)hypohalous acids and hydrogen peroxide dynamics in a zebrafish tail fin injury model.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Dröge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. - PubMed
-
- Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid. Redox Signal. 2008;10:1199–1234. - PubMed
-
- Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res Toxicol. 2001;14:1453–1464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

