Mutations in Hcfc1 and Ronin result in an inborn error of cobalamin metabolism and ribosomopathy
- PMID: 35013307
- PMCID: PMC8748873
- DOI: 10.1038/s41467-021-27759-7
Mutations in Hcfc1 and Ronin result in an inborn error of cobalamin metabolism and ribosomopathy
Abstract
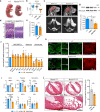
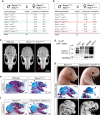
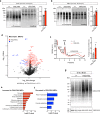
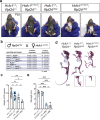
Combined methylmalonic acidemia and homocystinuria (cblC) is the most common inborn error of intracellular cobalamin metabolism and due to mutations in Methylmalonic Aciduria type C and Homocystinuria (MMACHC). Recently, mutations in the transcriptional regulators HCFC1 and RONIN (THAP11) were shown to result in cellular phenocopies of cblC. Since HCFC1/RONIN jointly regulate MMACHC, patients with mutations in these factors suffer from reduced MMACHC expression and exhibit a cblC-like disease. However, additional de-regulated genes and the resulting pathophysiology is unknown. Therefore, we have generated mouse models of this disease. In addition to exhibiting loss of Mmachc, metabolic perturbations, and developmental defects previously observed in cblC, we uncovered reduced expression of target genes that encode ribosome protein subunits. We also identified specific phenotypes that we ascribe to deregulation of ribosome biogenesis impacting normal translation during development. These findings identify HCFC1/RONIN as transcriptional regulators of ribosome biogenesis during development and their mutation results in complex syndromes exhibiting aspects of both cblC and ribosomopathies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Froese DS, Fowler B, Baumgartner MR. Vitamin B12, folate, and the methionine remethylation cycle-biochemistry, pathways, and regulation. J. Inherit. Metab. Dis. 2019;42:673–685. - PubMed
-
- Watkins D, Rosenblatt DS. Lessons in biology from patients with inborn errors of vitamin B12 metabolism. Biochimie. 2013;95:1019–1022. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

