The m6A methyltransferase METTL3 regulates muscle maintenance and growth in mice
- PMID: 35013323
- PMCID: PMC8748755
- DOI: 10.1038/s41467-021-27848-7
The m6A methyltransferase METTL3 regulates muscle maintenance and growth in mice
Abstract
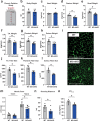
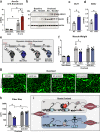
Skeletal muscle serves fundamental roles in organismal health. Gene expression fluctuations are critical for muscle homeostasis and the response to environmental insults. Yet, little is known about post-transcriptional mechanisms regulating such fluctuations while impacting muscle proteome. Here we report genome-wide analysis of mRNA methyladenosine (m6A) dynamics of skeletal muscle hypertrophic growth following overload-induced stress. We show that increases in METTL3 (the m6A enzyme), and concomitantly m6A, control skeletal muscle size during hypertrophy; exogenous delivery of METTL3 induces skeletal muscle growth, even without external triggers. We also show that METTL3 represses activin type 2 A receptors (ACVR2A) synthesis, blunting activation of anti-hypertrophic signaling. Notably, myofiber-specific conditional genetic deletion of METTL3 caused spontaneous muscle wasting over time and abrogated overload-induced hypertrophy; a phenotype reverted by co-administration of a myostatin inhibitor. These studies identify a previously unrecognized post-transcriptional mechanism promoting the hypertrophic response of skeletal muscle via control of myostatin signaling.
© 2022. The Author(s).
Conflict of interest statement
C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc. The remaining authors declare no competing interests.
Figures




References
-
- Lloyd AC. The regulation of cell size. Cell. 2013;154:1194–1205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

