Development and validation of a circulating microRNA panel for the early detection of breast cancer
- PMID: 35013577
- PMCID: PMC8810862
- DOI: 10.1038/s41416-021-01593-6
Development and validation of a circulating microRNA panel for the early detection of breast cancer
Erratum in
-
Correction: Development and validation of a circulating microRNA panel for the early detection of breast cancer.Br J Cancer. 2024 Oct;131(6):1107. doi: 10.1038/s41416-024-02800-w. Br J Cancer. 2024. PMID: 39191896 Free PMC article. No abstract available.
Abstract
Background: Mammography is widely used for breast cancer screening but suffers from a high false-positive rate. Here, we perform the largest comprehensive, multi-center study to date involving diverse ethnic groups, for the identification of circulating miRNAs for breast cancer screening.
Methods: This study had a discovery phase (n = 289) and two validation phases (n = 374 and n = 379). Quantitative PCR profiling of 324 miRNAs was performed on serum samples from breast cancer (all stages) and healthy subjects to identify miRNA biomarkers. Two-fold cross-validation was used for building and optimising breast cancer-associated miRNA panels. An optimal panel was validated in cohorts with Caucasian and Asian samples. Diagnostic ability was evaluated using area under the curve (AUC) analysis.
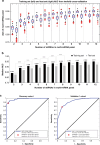
Results: The study identified and validated 30 miRNAs dysregulated in breast cancer. An optimised eight-miRNA panel showed consistent performance in all cohorts and was successfully validated with AUC, accuracy, sensitivity, and specificity of 0.915, 82.3%, 72.2% and 91.5%, respectively. The prediction model detected breast cancer in both Caucasian and Asian populations with AUCs ranging from 0.880 to 0.973, including pre-malignant lesions (stage 0; AUC of 0.831) and early-stage (stages I-II) cancers (AUC of 0.916).
Conclusions: Our panel can potentially be used for breast cancer screening, in conjunction with mammography.
© 2021. The Author(s).
Conflict of interest statement
HPT, LZ and RZ are shareholders of MiRXES Pte Ltd. LZ, RZ and YCT are employees of MiRXES Pte Ltd. All other authors declare no competing interests.
Figures



References
-
- Loke SY, Lee ASG. The future of blood-based biomarkers for the early detection of breast cancer. Eur J Cancer. 2018;92:54–68. - PubMed
-
- Ong MS, Mandl KD. National expenditure for false-positive mammograms and breast cancer overdiagnoses estimated at $4 billion a year. Health Aff (Millwood). 2015;34:576–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

