Sequence preference and scaffolding requirement for the inhibition of human neutrophil elastase by ecotin peptide
- PMID: 35014748
- PMCID: PMC8927871
- DOI: 10.1002/pro.4274
Sequence preference and scaffolding requirement for the inhibition of human neutrophil elastase by ecotin peptide
Abstract
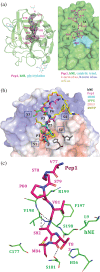
Human neutrophil elastase (hNE) is an abundant serine protease that is a major constituent of lung elastolytic activity. However, when secreted in excess, if not properly attenuated by selective inhibitor proteins, it can have detrimental effects on host tissues, leading to chronic lung inflammation and non-small cell lung cancer. To improve upon the design of inhibitors against hNE for therapeutic applications, here, we report the crystal structure of hNE in complex with an ecotin (ET)-derived peptide inhibitor. We show that the peptide binds in the nonprime substrate binding site. Unexpectedly, compared with full-length (FL) ET, we find that our short linear peptides and circular amide backbone-linked peptides of ET are incapable of efficient hNE inhibition. Our structural insights point to a preferred amino acid sequence and the potential benefit of a scaffold for optimal binding and function of the peptide inhibitor, both of which are retained in the FL ET protein. These findings will aid in the development of effective peptide-based inhibitors against hNE for targeted therapy.
Keywords: elastase; inhibition; peptide; serine protease; structure.
© 2022 The Protein Society.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


Similar articles
-
Structural Basis for the Inhibition Mechanism of Ecotin against Neutrophil Elastase by Targeting the Active Site and Secondary Binding Site.Biochemistry. 2020 Aug 4;59(30):2788-2795. doi: 10.1021/acs.biochem.0c00493. Epub 2020 Jul 21. Biochemistry. 2020. PMID: 32657577
-
Discriminating between the activities of human neutrophil elastase and proteinase 3 using serpin-derived fluorogenic substrates.J Biol Chem. 2002 Oct 18;277(42):39074-81. doi: 10.1074/jbc.M202918200. Epub 2002 Jul 11. J Biol Chem. 2002. PMID: 12114510
-
The periplasmic serine protease inhibitor ecotin protects bacteria against neutrophil elastase.Biochem J. 2004 Apr 1;379(Pt 1):107-18. doi: 10.1042/BJ20031790. Biochem J. 2004. PMID: 14705961 Free PMC article.
-
A patenting perspective on human neutrophil elastase (HNE) inhibitors (2014-2018) and their therapeutic applications.Expert Opin Ther Pat. 2019 Jul;29(7):555-578. doi: 10.1080/13543776.2019.1630379. Epub 2019 Jun 16. Expert Opin Ther Pat. 2019. PMID: 31204543 Free PMC article. Review.
-
Neutrophil elastase as a target in lung cancer.Anticancer Agents Med Chem. 2012 Jul;12(6):565-79. doi: 10.2174/187152012800617696. Anticancer Agents Med Chem. 2012. PMID: 22263788 Review.
References
-
- Stockley R, De Soyza A, Gunawardena K, et al. Phase II study of a neutrophil elastase inhibitor (AZD9668) in patients with bronchiectasis. Respir Med. 2013;107:524–533. - PubMed
-
- Von Nussbaum F, Li M‐J. Neutrophil elastase inhibitors for the treatment of (cardio) pulmonary diseases: Into clinical testing with pre‐adaptive pharmacophores. Bioorg Med Chem Lett. 2015;25:4370–4381. - PubMed
-
- Zeiher BG, Matsuoka S, Kawabata K, Repine JE. Neutrophil elastase and acute lung injury: Prospects for sivelestat and other neutrophil elastase inhibitors as therapeutics. Crit Care Med. 2002;30:S281–S287. - PubMed
-
- Yamashita J, Ogawa M, Abe M, et al. Tumor neutrophil elastase is closely associated with the direct extension of non‐small cell lung cancer into the aorta. Chest. 1997;111:885–890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

