Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015
- PMID: 35014866
- PMCID: PMC8751382
- DOI: 10.1128/msystems.01169-21
Specificities and Commonalities of Carbapenemase-Producing Escherichia coli Isolated in France from 2012 to 2015
Abstract
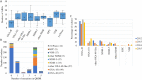
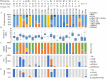
Carbapenemase-producing Escherichia coli (CP-Ec) represents a major public health threat with a risk of dissemination in the community as has occurred for lineages producing extended-spectrum β-lactamases. To characterize the extent of CP-Ec spread in France, isolates from screening and infection samples received at the French National Reference Center (F-NRC) laboratory for carbapenemase-producing Enterobacterales were investigated. A total of 691 CP-Ec isolates collected between 2012 and 2015 and 22 isolates collected before 2012 were fully sequenced. Analysis of their genome sequences revealed some disseminating multidrug-resistant (MDR) lineages frequently acquiring diverse carbapenemase genes mainly belonging to clonal complex 23 (CC23) (sequence type 410 [ST410]) and CC10 (ST10 and ST167) and sporadic isolates, including rare ST131 isolates (n = 17). However, the most represented sequence type (ST) was ST38 (n = 92) with four disseminated lineages carrying blaOXA-48-like genes inserted in the chromosome. Globally, the most frequent carbapenemase gene (n = 457) was blaOXA-48. It was also less frequently associated with MDR isolates being the only resistance gene in 119 isolates. Thus, outside the ST38 clades, its acquisition was frequently sporadic with no sign of dissemination, reflecting the circulation of the IncL plasmid pOXA-48 in France and its high frequency of conjugation. In contrast, blaOXA-181 and blaNDM genes were often associated with the evolution of MDR E. coli lineages characterized by mutations in ftsI and ompC. IMPORTANCE Carbapenemase-producing Escherichia coli (CP-Ec) might be difficult to detect, as MICs can be very low. However, their absolute number and their proportion among carbapenem-resistant Enterobacterales have been increasing, as reported by WHO and national surveillance programs. This suggests a still largely uncharacterized community spread of these isolates. Here, we have characterized the diversity and evolution of CP-Ec isolated in France before 2016. We show that carbapenemase genes are associated with a wide variety of E. coli genomic backgrounds and a small number of dominant phylogenetic lineages. In a significant proportion of CP-Ec, the most frequent carbapenemase gene blaOXA-48, was detected in isolates lacking any other resistance gene, reflecting the dissemination of pOXA-48 plasmids, likely in the absence of any antibiotic pressure. In contrast, carbapenemase gene transfer may also occur in multidrug-resistant E. coli, ultimately giving rise to at-risk lineages encoding carbapenemases with a high potential of dissemination.
Keywords: drug resistance evolution; emergence; genomics; multidrug resistance; surveillance studies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- French National Reference Center for antibiotic resistance, Le Kremlin-Bicêtre, France, activity report 2019–2020. https://www.cnr-resistance-antibiotiques.fr/bilans-dactivite.html.
-
- Saleem AF, Allana A, Hale L, Diaz A, Salinas R, Salinas C, Qureshi SM, Hotwani A, Rahman N, Khan A, Zaidi AK, Seed PC, Arshad M. 2020. The gut of healthy infants in the community as a reservoir of ESBL and carbapenemase-producing bacteria. Antibiotics 9:286. doi: 10.3390/antibiotics9060286. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
