Quiescent human glioblastoma cancer stem cells drive tumor initiation, expansion, and recurrence following chemotherapy
- PMID: 35016005
- PMCID: PMC8820651
- DOI: 10.1016/j.devcel.2021.12.007
Quiescent human glioblastoma cancer stem cells drive tumor initiation, expansion, and recurrence following chemotherapy
Abstract
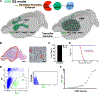
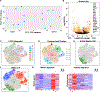
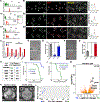
We test the hypothesis that glioblastoma harbors quiescent cancer stem cells that evade anti-proliferative therapies. Functional characterization of spontaneous glioblastomas from genetically engineered mice reveals essential quiescent stem-like cells that can be directly isolated from tumors. A derived quiescent cancer-stem-cell-specific gene expression signature is enriched in pre-formed patient GBM xenograft single-cell clusters that lack proliferative gene expression. A refined human 118-gene signature is preserved in quiescent single-cell populations from primary and recurrent human glioblastomas. The F3 cell-surface receptor mRNA, expressed in the conserved signature, identifies quiescent tumor cells by antibody immunohistochemistry. F3-antibody-sorted glioblastoma cells exhibit stem cell gene expression, enhance self-renewal in culture, drive tumor initiation and serial transplantation, and reconstitute tumor heterogeneity. Upon chemotherapy, the spared cancer stem cell pool becomes activated and accelerates transition to proliferation. These results help explain conventional treatment failure and lay a conceptual framework for alternative therapies.
Keywords: F3 receptor; cancer stem cells; chemoresistance; glioblastoma; heterogeneity; quiescence; recurrence; single-cell RNA sequencing; temozolomide.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Alvarez-Satta M, Moreno-Cugnon L, and Matheu A (2019). Primary cilium and brain aging: role in neural stem cells, neurodegenerative diseases and glioblastoma. Ageing Res Rev 52, 53–63. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

