LTK fusions: A new target emerges in non-small cell lung cancer
- PMID: 35016028
- PMCID: PMC9574472
- DOI: 10.1016/j.ccell.2021.12.012
LTK fusions: A new target emerges in non-small cell lung cancer
Abstract
Identification of targetable fusions as oncogenic drivers in non-small cell lung cancer has transformed its diagnostic and therapeutic paradigm. In a recent article in Nature, Izumi et al. report the discovery of CLIP1-LTK fusion as a novel oncogenic driver in lung cancer, targetable using the ALK tyrosine kinase inhibitor lorlatinib.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.J.C. declares no competing interests. L.V.S. has received consulting fees from AstraZeneca, Genentech, Pfizer, Takeda, and Janssen and has received institutional research support from Boehringer-Ingelheim, Novartis, AstraZeneca, and Delfi. T.W.J. is an employee of Pfizer. J.J.L. has served as a compensated consultant for Genentech, C4 Therapeutics, Blueprint Medicines, Nuvalent, and Turning Point Therapeutics; received honorarium and travel support from Pfizer; received institutional research funds from Hengrui Therapeutics, Turning Point Therapeutics, Neon Therapeutics, Relay Therapeutics, Bayer, Elevation Oncology, Roche, Linnaeus, and Novartis; and received CME funding from OncLive, MedStar Health, and Northwell Health.
Figures
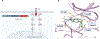
Comment on
-
The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer.Nature. 2021 Dec;600(7888):319-323. doi: 10.1038/s41586-021-04135-5. Epub 2021 Nov 24. Nature. 2021. PMID: 34819663 Free PMC article.
References
-
- Ben-Neriah Y, and Bauskin AR (1988). Leukocytes express a novel gene encoding a putative transmembrane protein-kinase devoid of an extracellular domain. Nature 333, 672–676. - PubMed
-
- Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, Ptashkin R, Won H, Chang J, Halpenny D, et al. (2019). High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res 25, 4712–4722. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

