Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization
- PMID: 35016196
- PMCID: PMC8866126
- DOI: 10.1038/s41586-021-04387-1
Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization
Abstract
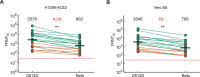
The emergence of the SARS-CoV-2 variant of concern Omicron (Pango lineage B.1.1.529), first identified in Botswana and South Africa, may compromise vaccine effectiveness and lead to re-infections1. Here we investigated Omicron escape from neutralization by antibodies from South African individuals vaccinated with Pfizer BNT162b2. We used blood samples taken soon after vaccination from individuals who were vaccinated and previously infected with SARS-CoV-2 or vaccinated with no evidence of previous infection. We isolated and sequence-confirmed live Omicron virus from an infected person and observed that Omicron requires the angiotensin-converting enzyme 2 (ACE2) receptor to infect cells. We compared plasma neutralization of Omicron relative to an ancestral SARS-CoV-2 strain and found that neutralization of ancestral virus was much higher in infected and vaccinated individuals compared with the vaccinated-only participants. However, both groups showed a 22-fold reduction in vaccine-elicited neutralization by the Omicron variant. Participants who were vaccinated and had previously been infected exhibited residual neutralization of Omicron similar to the level of neutralization of the ancestral virus observed in the vaccination-only group. These data support the notion that reasonable protection against Omicron may be maintained using vaccination approaches.
© 2021. The Author(s).
Conflict of interest statement
Salim S. Abdool Karim is a member of the COVID advisory panel for emerging markets at Pfizer. The authors declare no other competing interests.
Figures

Update of
-
SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection.medRxiv [Preprint]. 2021 Dec 17:2021.12.08.21267417. doi: 10.1101/2021.12.08.21267417. medRxiv. 2021. Update in: Nature. 2022 Feb;602(7898):654-656. doi: 10.1038/s41586-021-04387-1. PMID: 34909788 Free PMC article. Updated. Preprint.
-
Omicron infection of vaccinated individuals enhances neutralizing immunity against the Delta variant.medRxiv [Preprint]. 2022 Jan 28:2021.12.27.21268439. doi: 10.1101/2021.12.27.21268439. medRxiv. 2022. Update in: Nature. 2022 Feb;602(7898):654-656. doi: 10.1038/s41586-021-04387-1. Update in: Nature. 2022 Jul;607(7918):356-359. doi: 10.1038/s41586-022-04830-x. PMID: 34981076 Free PMC article. Updated. Preprint.
References
-
- Pulliam, J. R. C. et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. Preprint at 10.1101/2021.11.11.21266068 (2021).
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

