Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism
- PMID: 35017197
- PMCID: PMC9664128
- DOI: 10.1136/gutjnl-2021-325753
Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism
Abstract
Objectives: Gut microbiota is a key component in obesity and type 2 diabetes, yet mechanisms and metabolites central to this interaction remain unclear. We examined the human gut microbiome's functional composition in healthy metabolic state and the most severe states of obesity and type 2 diabetes within the MetaCardis cohort. We focused on the role of B vitamins and B7/B8 biotin for regulation of host metabolic state, as these vitamins influence both microbial function and host metabolism and inflammation.
Design: We performed metagenomic analyses in 1545 subjects from the MetaCardis cohorts and different murine experiments, including germ-free and antibiotic treated animals, faecal microbiota transfer, bariatric surgery and supplementation with biotin and prebiotics in mice.
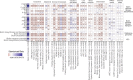
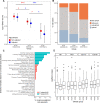
Results: Severe obesity is associated with an absolute deficiency in bacterial biotin producers and transporters, whose abundances correlate with host metabolic and inflammatory phenotypes. We found suboptimal circulating biotin levels in severe obesity and altered expression of biotin-associated genes in human adipose tissue. In mice, the absence or depletion of gut microbiota by antibiotics confirmed the microbial contribution to host biotin levels. Bariatric surgery, which improves metabolism and inflammation, associates with increased bacterial biotin producers and improved host systemic biotin in humans and mice. Finally, supplementing high-fat diet-fed mice with fructo-oligosaccharides and biotin improves not only the microbiome diversity, but also the potential of bacterial production of biotin and B vitamins, while limiting weight gain and glycaemic deterioration.
Conclusion: Strategies combining biotin and prebiotic supplementation could help prevent the deterioration of metabolic states in severe obesity.
Trial registration number: NCT02059538.
Keywords: diabetes mellitus; intestinal bacteria; micronutrients; nutrition; obesity.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: KC is a consultant for Danone Research, Ysopia and CONFO therapeutics for work not associated with this study. KC held a collaborative research contract with Danone Research in the context of MetaCardis project. FB is a shareholder of Implexion pharma AB. MB received lecture and/or consultancy fees from AstraZeneca, Boehringer-Ingelheim, Lilly, Novo Nordisk, Novartis and Sanofi.
Figures


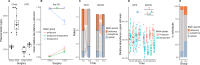

Comment in
-
Tiny contributors to severe obesity inside the gut.Gut. 2022 Dec;71(12):2376-2378. doi: 10.1136/gutjnl-2021-326781. Epub 2022 Feb 8. Gut. 2022. PMID: 35135844 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
