A BAFF ligand-based CAR-T cell targeting three receptors and multiple B cell cancers
- PMID: 35017485
- PMCID: PMC8752722
- DOI: 10.1038/s41467-021-27853-w
A BAFF ligand-based CAR-T cell targeting three receptors and multiple B cell cancers
Abstract
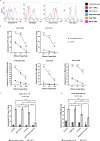
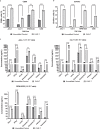
B cell-activating factor (BAFF) binds the three receptors BAFF-R, BCMA, and TACI, predominantly expressed on mature B cells. Almost all B cell cancers are reported to express at least one of these receptors. Here we develop a BAFF ligand-based chimeric antigen receptor (CAR) and generate BAFF CAR-T cells using a non-viral gene delivery method. We show that BAFF CAR-T cells bind specifically to each of the three BAFF receptors and are effective at killing multiple B cell cancers, including mantle cell lymphoma (MCL), multiple myeloma (MM), and acute lymphoblastic leukemia (ALL), in vitro and in vivo using different xenograft models. Co-culture of BAFF CAR-T cells with these tumor cells results in induction of activation marker CD69, degranulation marker CD107a, and multiple proinflammatory cytokines. In summary, we report a ligand-based BAFF CAR-T capable of binding three different receptors, minimizing the potential for antigen escape in the treatment of B cell cancers.
© 2022. The Author(s).
Conflict of interest statement
N.S. and S.D. are employees of Luminary Therapeutics. R.P. is a consultant for Luminary therapeutics. B.J.J. is an employee at Bio-Techne. B.R.W. and B.S.M. are consultants for Bio-Techne and are shareholders of Luminary Therapeutics. P.C. is a member of advisory board of ADC therapeutics and have received honoraria from kite pharma. The remaining authors declare no competing interests.
Figures





References
-
- Novak AJ, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

