Canonical WNT signaling-dependent gating of MYC requires a noncanonical CTCF function at a distal binding site
- PMID: 35017527
- PMCID: PMC8752836
- DOI: 10.1038/s41467-021-27868-3
Canonical WNT signaling-dependent gating of MYC requires a noncanonical CTCF function at a distal binding site
Abstract
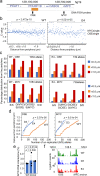
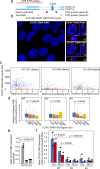
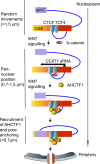
Abnormal WNT signaling increases MYC expression in colon cancer cells in part via oncogenic super-enhancer-(OSE)-mediated gating of the active MYC to the nuclear pore in a poorly understood process. We show here that the principal tenet of the WNT-regulated MYC gating, facilitating nuclear export of the MYC mRNA, is regulated by a CTCF binding site (CTCFBS) within the OSE to confer growth advantage in HCT-116 cells. To achieve this, the CTCFBS directs the WNT-dependent trafficking of the OSE to the nuclear pore from intra-nucleoplasmic positions in a stepwise manner. Once the OSE reaches a peripheral position, which is triggered by a CTCFBS-mediated CCAT1 eRNA activation, its final stretch (≤0.7 μm) to the nuclear pore requires the recruitment of AHCTF1, a key nucleoporin, to the CTCFBS. Thus, a WNT/ß-catenin-AHCTF1-CTCF-eRNA circuit enables the OSE to promote pathological cell growth by coordinating the trafficking of the active MYC gene within the 3D nuclear architecture.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

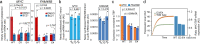


References
-
- Cremer T, et al. The 4D nucleome: evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–2943. - PubMed
-
- Pascual-Garcia P, Capelson M. Nuclear pores as versatile platforms for gene regulation. Curr. Opin. Genet. Dev. 2014;25:110–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

