Establishment and validation of an autophagy-related prognostic signature for survival predicting in cutaneous melanoma
- PMID: 35018237
- PMCID: PMC8727816
Establishment and validation of an autophagy-related prognostic signature for survival predicting in cutaneous melanoma
Abstract
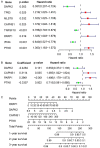
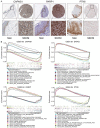
Existing staging system for prognosis evaluating for Skin Cutaneous Melanoma (SKCM) patients had defects of subjective, inaccuracy and inconsistently, therefore, to identify specific and applicable prognostic markers and promote personalized therapeutic interventions is urgently required. This study aims to build a robust autophagy-related genes (ARGs) signature for prognosis monitoring of SKCM patients. We determined 26 ARGs as differentially expressed autophagy-related genes (DEARGs) from 103 SKCM and 23 normal skin samples in GSE15605 and GSE3189 datasets. Optimal prognostic DEARGs composed the risk model were screened and verified in 458 SKCM patients in TCGA cohort as the training cohort and 209 patients in GSE65904 as the test cohort. Finally, 4 optimal independent prognostic DEARGs (CAPNS1, DAPK2, PARP1 and PTK6) were filtered out in the training cohort to establish the risk model. A prognostic nomogram was established for quantitative survival prediction. The risk model grouped high-risk SKCM cancer patients exhibited significantly shorter survival times in both training and test cohorts. The area under the ROC curve for risk score model was 0.788 and 0.627 in the training and test cohorts indicated the risk model was relatively accurate for prognosis monitoring. Clinical correlation analysis exhibited that risk score was an independent predictor for prognosis significantly associated with T/N classification. The prognostic value of the 4 risk genes formed the risk model was also validated respectively. We identified a novel autophagy-related signature for prognosis monitoring. It has the potential to be an independent prognostic indicator and can benefit targeted therapy.
Keywords: Skin cutaneous melanoma; autophagy-related genes; prognostic risk model; survival prediction; targeted therapeutic intervention.
AJCR Copyright © 2021.
Conflict of interest statement
None.
Figures





References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A, Ugurel S. Melanoma. Lancet. 2018;392:971–984. - PubMed
-
- Garbe C, Peris K, Hauschild A, Saiag P, Middleton M, Spatz A, Grob JJ, Malvehy J, Newton-Bishop J, Stratigos A, Pehamberger H, Eggermont AM European Dermatology Forum, European Association of Dermato-Oncology, European Organization of Research and Treatment of Cancer. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline--update 2012. Eur J Cancer. 2012;48:2375–2390. - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. - PubMed
-
- McKinnon JG, Yu XQ, McCarthy WH, Thompson JF. Prognosis for patients with thin cutaneous melanoma: long-term survival data from New South Wales Central Cancer Registry and the Sydney Melanoma Unit. Cancer. 2003;98:1223–1231. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
