Comprehensive analysis of prime editing outcomes in human embryonic stem cells
- PMID: 35018468
- PMCID: PMC8789035
- DOI: 10.1093/nar/gkab1295
Comprehensive analysis of prime editing outcomes in human embryonic stem cells
Abstract
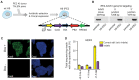
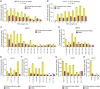
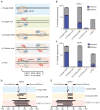
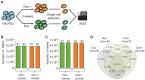
Prime editing is a versatile and precise genome editing technique that can directly copy desired genetic modifications into target DNA sites without the need for donor DNA. This technique holds great promise for the analysis of gene function, disease modeling, and the correction of pathogenic mutations in clinically relevant cells such as human pluripotent stem cells (hPSCs). Here, we comprehensively tested prime editing in hPSCs by generating a doxycycline-inducible prime editing platform. Prime editing successfully induced all types of nucleotide substitutions and small insertions and deletions, similar to observations in other human cell types. Moreover, we compared prime editing and base editing for correcting a disease-related mutation in induced pluripotent stem cells derived form a patient with α 1-antitrypsin (A1AT) deficiency. Finally, whole-genome sequencing showed that, unlike the cytidine deaminase domain of cytosine base editors, the reverse transcriptase domain of a prime editor does not lead to guide RNA-independent off-target mutations in the genome. Our results demonstrate that prime editing in hPSCs has great potential for complementing previously developed CRISPR genome editing tools.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hotta A., Yamanaka S.. From genomics to gene therapy: induced pluripotent stem cells meet genome editing. Annu. Rev. Genet. 2015; 49:47–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

