Mutations at the Alphavirus E1'-E2 Interdimer Interface Have Host-Specific Phenotypes
- PMID: 35019719
- PMCID: PMC8906394
- DOI: 10.1128/jvi.02149-21
Mutations at the Alphavirus E1'-E2 Interdimer Interface Have Host-Specific Phenotypes
Abstract
Alphaviruses are enveloped viruses transmitted by arthropod vectors to vertebrate hosts. The surface of the virion contains 80 glycoprotein spikes embedded in the membrane, and these spikes mediate attachment to the host cell and initiate viral fusion. Each spike consists of a trimer of E2-E1 heterodimers. These heterodimers interact at the following two interfaces: (i) the intradimer interactions between E2 and E1 of the same heterodimer and (ii) the interdimer interactions between E2 of one heterodimer and E1 of the adjacent heterodimer (E1'). We hypothesized that the interdimer interactions are essential for trimerization of the E2-E1 heterodimers into a functional spike. In this work, we made a mutant virus (chikungunya piggyback [CPB]) where we replaced six interdimeric residues in the E2 protein of Sindbis virus (wild-type [WT] SINV) with those from the E2 protein from chikungunya virus and studied its effect in both mammalian and mosquito cell lines. CPB produced fewer infectious particles in mammalian cells than in mosquito cells, relative to WT SINV. When CPB virus was purified from mammalian cells, particles showed reduced amounts of glycoproteins relative to the capsid protein and contained defects in particle morphology compared with virus derived from mosquito cells. Using cryo-electron microscopy (cryo-EM), we determined that the spikes of CPB had a different conformation than WT SINV. Last, we identified two revertants, E2-H333N and E1-S247L, that restored particle growth and assembly to different degrees. We conclude the interdimer interface is critical for spike trimerization and is a novel target for potential antiviral drug design. IMPORTANCE Alphaviruses, which can cause disease when spread to humans by mosquitoes, have been classified as emerging pathogens, with infections occurring worldwide. The spikes on the surface of the alphavirus particle are absolutely required for the virus to enter a new host cell and initiate an infection. Using a structure-guided approach, we made a mutant virus that alters spike assembly in mammalian cells but not mosquito cells. This finding is important because it identifies a region in the spike that could be a target for antiviral drug design.
Keywords: Glycoprotein spike; alphaviruses; assembly; host-range.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

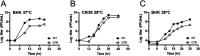

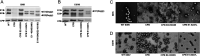

References
-
- Griffin DE. 2013. Chapter 23: alphaviruses, p 652–770. In Knipe DM, Howley PM (ed), Fields’ virology. Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Kuhn RJ. 2013. Chapter 22: Togaviradae, p 629–650. In Knipe DM, Howley PM (ed), Fields’ virology. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

