Rapid SARS-CoV-2 Adaptation to Available Cellular Proteases
- PMID: 35019723
- PMCID: PMC8906416
- DOI: 10.1128/jvi.02186-21
Rapid SARS-CoV-2 Adaptation to Available Cellular Proteases
Abstract
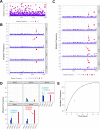
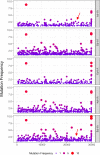
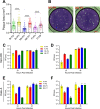
Recent emergence of SARS-CoV-1 variants demonstrates the potential of this virus for targeted evolution, despite its overall genomic stability. Here we show the dynamics and the mechanisms behind the rapid adaptation of SARS-CoV-2 to growth in Vero E6 cells. The selective advantage for growth in Vero E6 cells is due to increased cleavage efficiency by cathepsins at the mutated S1/S2 site. S1/S2 site also constitutes a heparan sulfate (HS) binding motif that influenced virus growth in Vero E6 cells, but HS antagonist did not inhibit virus adaptation in these cells. The entry of Vero E6-adapted virus into human cells is defective because the mutated spike variants are poorly processed by furin or TMPRSS2. Minor subpopulation that lack the furin cleavage motif in the spike protein rapidly become dominant upon passaging through Vero E6 cells, but wild type sequences are maintained at low percentage in the virus swarm and mediate a rapid reverse adaptation if the virus is passaged again on TMPRSS2+ human cells. Our data show that the spike protein of SARS-CoV-2 can rapidly adapt itself to available proteases and argue for deep sequence surveillance to identify the emergence of novel variants. IMPORTANCE Recently emerging SARS-CoV-2 variants B.1.1.7 (alpha variant), B.1.617.2 (delta variant), and B.1.1.529 (omicron variant) harbor spike mutations and have been linked to increased virus pathogenesis. The emergence of these novel variants highlights coronavirus adaptation and evolution potential, despite the stable consensus genotype of clinical isolates. We show that subdominant variants maintained in the virus population enable the virus to rapidly adapt to selection pressure. Although these adaptations lead to genotype change, the change is not absolute and genomes with original genotype are maintained in the virus swarm. Thus, our results imply that the relative stability of SARS-CoV-2 in numerous independent clinical isolates belies its potential for rapid adaptation to new conditions.
Keywords: SARS-CoV-2; coronavirus spike priming; deep sequencing; furin cleavage site; spike mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

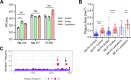

References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, Research T . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

