Microbiome Resilience despite a Profound Loss of Minority Microbiota following Clindamycin Challenge in Humanized Gnotobiotic Mice
- PMID: 35019780
- PMCID: PMC8754120
- DOI: 10.1128/spectrum.01960-21
Microbiome Resilience despite a Profound Loss of Minority Microbiota following Clindamycin Challenge in Humanized Gnotobiotic Mice
Abstract
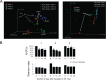
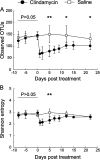
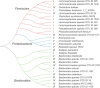
Antibiotics are known to induce gut dysbiosis and increase the risk of antibiotic resistance. While antibiotic exposure is a known risk factor leading to compromised colonization resistance against enteric pathogens such as Clostridioides difficile, the extent and consequences of antibiotic perturbation on the human gut microbiome remain poorly understood. Human studies on impacts of antibiotics are complicated by the tremendous variability of gut microbiome among individuals, even between identical twins. Furthermore, antibiotic challenge experiments cannot be replicated in human subjects for a given gut microbiome. Here, we transplanted feces from three unrelated human donors into groups of identical germfree (GF) Swiss-Webster mice, and examined the temporal responses of the transplanted microbiome to oral clindamycin challenge in gnotobiotic isolators over 7 weeks. Analysis of 177 longitudinal fecal samples revealed that 59% to 81% of human microbiota established a stable configuration rapidly and stably in GF mice. Microbiome responses to clindamycin challenge was highly reproducible and microbiome-dependent. A short course of clindamycin was sufficient to induce a profound loss (∼one third) of the microbiota by disproportionally eliminating minority members of the transplanted microbiota. However, it was inadequate to disrupt the global microbial community structure or function, which rebounded rapidly to resemble its pre-treatment state after clindamycin discontinuation. Furthermore, the response of individual microbes was community-dependent. Taken together, these results suggest that the overall gut microbiome structure is resilient to antibiotic perturbation, the functional consequences of which warrant further investigation. IMPORTANCE Antibiotics cause imbalance of gut microbiota, which in turn increase our susceptibility to gastrointestinal infections. However, how antibiotics disrupt gut bacterial communities is not well understood, and exposing healthy volunteers to unnecessary antibiotics for research purposes carries clinical and ethical concerns. In this study, we used genetically identical mice transplanted with the same human gut microbiota to control for both genetic and environmental variables. We found that a short course of oral clindamycin was sufficient to eliminate one third of the gut bacteria by disproportionally eliminating minority members of the transplanted microbiota, but it was inadequate to disrupt the overall microbial community structure and function, which rebounded rapidly to its pre-treatment state. These results suggest that gut microbiome is highly resilient to antibiotic challenge and degradation of the human gut ecosystem may require repeated or prolonged antibiotic exposure.
Keywords: clindamycin; gut dysbiosis; humanized gnotobiotic mice; microbiome; resilience.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

