Analysis of HubP-dependent cell pole protein targeting in Vibrio cholerae uncovers novel motility regulators
- PMID: 35020734
- PMCID: PMC8789113
- DOI: 10.1371/journal.pgen.1009991
Analysis of HubP-dependent cell pole protein targeting in Vibrio cholerae uncovers novel motility regulators
Erratum in
-
Correction: Analysis of HubP-dependent cell pole protein targeting in Vibrio cholerae uncovers novel motility regulators.PLoS Genet. 2022 Mar 1;18(3):e1010094. doi: 10.1371/journal.pgen.1010094. eCollection 2022 Mar. PLoS Genet. 2022. PMID: 35231028 Free PMC article.
Abstract
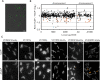
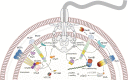
In rod-shaped bacteria, the emergence and maintenance of long-axis cell polarity is involved in key cellular processes such as cell cycle, division, environmental sensing and flagellar motility among others. Many bacteria achieve cell pole differentiation through the use of polar landmark proteins acting as scaffolds for the recruitment of functional macromolecular assemblies. In Vibrio cholerae a large membrane-tethered protein, HubP, specifically interacts with proteins involved in chromosome segregation, chemotaxis and flagellar biosynthesis. Here we used comparative proteomics, genetic and imaging approaches to identify additional HubP partners and demonstrate that at least six more proteins are subject to HubP-dependent polar localization. These include a cell-wall remodeling enzyme (DacB), a likely chemotaxis sensory protein (HlyB), two presumably cytosolic proteins of unknown function (VC1210 and VC1380) and two membrane-bound proteins, named here MotV and MotW, that exhibit distinct effects on chemotactic motility. We show that while both ΔmotW and ΔmotV mutants retain monotrichous flagellation, they present significant to severe motility defects when grown in soft agar. Video-tracking experiments further reveal that ΔmotV cells can swim in liquid environments but are unable to tumble or penetrate a semisolid matrix, whereas a motW deletion affects both tumbling frequency and swimming speed. Motility suppressors and gene co-occurrence analyses reveal co-evolutionary linkages between MotV, a subset of non-canonical CheV proteins and flagellar C-ring components FliG and FliM, whereas MotW regulatory inputs appear to intersect with specific c-di-GMP signaling pathways. Together, these results reveal an ever more versatile role for the landmark cell pole organizer HubP and identify novel mechanisms of motility regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

