BioThings SDK: a toolkit for building high-performance data APIs in biomedical research
- PMID: 35020801
- PMCID: PMC8963279
- DOI: 10.1093/bioinformatics/btac017
BioThings SDK: a toolkit for building high-performance data APIs in biomedical research
Abstract
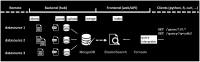
Summary: To meet the increased need of making biomedical resources more accessible and reusable, Web Application Programming Interfaces (APIs) or web services have become a common way to disseminate knowledge sources. The BioThings APIs are a collection of high-performance, scalable, annotation as a service APIs that automate the integration of biological annotations from disparate data sources. This collection of APIs currently includes MyGene.info, MyVariant.info and MyChem.info for integrating annotations on genes, variants and chemical compounds, respectively. These APIs are used by both individual researchers and application developers to simplify the process of annotation retrieval and identifier mapping. Here, we describe the BioThings Software Development Kit (SDK), a generalizable and reusable toolkit for integrating data from multiple disparate data sources and creating high-performance APIs. This toolkit allows users to easily create their own BioThings APIs for any data type of interest to them, as well as keep APIs up-to-date with their underlying data sources.
Availability and implementation: The BioThings SDK is built in Python and released via PyPI (https://pypi.org/project/biothings/). Its source code is hosted at its github repository (https://github.com/biothings/biothings.api).
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures
References
-
- Elastic.co. (2021) Scalability and resilience: clusters, nodes, and shards | Elasticsearch Guide [7.15] | Elastic. https://www.elastic.co/guide/en/elasticsearch/reference/current/scalabil... (29 November 2021, date last accessed).
-
- European Organization for Nuclear Research. (2013) Zenodo. CERN. https://doi.org/10.25495/7gxk-rd71.
-
- Lelong S. et al. (2015) MyChem.info | Chemical and Drug Annotation as a Service. MyChem.info. http://mychem.info/ (3 August 2021, date last accessed).
-
- Lelong S. et al. (2017) BioThings Taxonomy API. https://t.biothings.io/ (1 October 2021, date last accessed).
-
- Lelong S. (2020a) MVCGI BioThings Studio data plugin demo. https://github.com/sirloon/mvcgi (1 October 2021, date last accessed).