ProteinBERT: a universal deep-learning model of protein sequence and function
- PMID: 35020807
- PMCID: PMC9386727
- DOI: 10.1093/bioinformatics/btac020
ProteinBERT: a universal deep-learning model of protein sequence and function
Abstract
Summary: Self-supervised deep language modeling has shown unprecedented success across natural language tasks, and has recently been repurposed to biological sequences. However, existing models and pretraining methods are designed and optimized for text analysis. We introduce ProteinBERT, a deep language model specifically designed for proteins. Our pretraining scheme combines language modeling with a novel task of Gene Ontology (GO) annotation prediction. We introduce novel architectural elements that make the model highly efficient and flexible to long sequences. The architecture of ProteinBERT consists of both local and global representations, allowing end-to-end processing of these types of inputs and outputs. ProteinBERT obtains near state-of-the-art performance, and sometimes exceeds it, on multiple benchmarks covering diverse protein properties (including protein structure, post-translational modifications and biophysical attributes), despite using a far smaller and faster model than competing deep-learning methods. Overall, ProteinBERT provides an efficient framework for rapidly training protein predictors, even with limited labeled data.
Availability and implementation: Code and pretrained model weights are available at https://github.com/nadavbra/protein_bert.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures


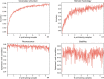

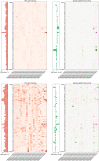
References
-
- Abadi M. et al. (2016) Tensorflow: a system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI16). pp. 265–283.
-
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

