Metabolic adaptation of lymphocytes in immunity and disease
- PMID: 35021054
- PMCID: PMC8842882
- DOI: 10.1016/j.immuni.2021.12.012
Metabolic adaptation of lymphocytes in immunity and disease
Abstract
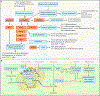
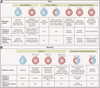
Adaptive immune responses mediated by T cells and B cells are crucial for protective immunity against pathogens and tumors. Differentiation and function of immune cells require dynamic reprogramming of cellular metabolism. Metabolic inputs, pathways, and enzymes display remarkable flexibility and heterogeneity, especially in vivo. How metabolic plasticity and adaptation dictate functional specialization of immune cells is fundamental to our understanding and therapeutic modulation of the immune system. Extensive progress has been made in characterizing the effects of metabolic networks on immune cell fate and function in discrete microenvironments or immunological contexts. In this review, we summarize how rewiring of cellular metabolism determines the outcome of adaptive immunity in vivo, with a focus on how metabolites, nutrients, and driver genes in immunometabolism instruct cellular programming and immune responses during infection, inflammation, and cancer in mice and humans. Understanding context-dependent metabolic remodeling will manifest legitimate opportunities for therapeutic intervention of human disease.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.C. is a consultant for Kumquat Biosciences.
Figures

References
-
- Angin M, Volant S, Passaes C, Lecuroux C, Monceaux V, Dillies MA, Valle-Casuso JC, Pancino G, Vaslin B, Le Grand R, et al. (2019). Metabolic plasticity of HIV-specific CD8(+) T cells is associated with enhanced antiviral potential and natural control of HIV-1 infection. Nat Metab 1, 704–716. - PubMed
-
- Arthur L, Esaulova E, Mogilenko DA, Tsurinov P, Burdess S, Laha A, Presti R, Goetz B, Watson MA, Goss CW, et al. (2021). Cellular and plasma proteomic determinants of COVID-19 and non-COVID-19 pulmonary diseases relative to healthy aging. Nature Aging 1, 535–549. - PubMed
-
- Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dahling S, Kastenmuller W, et al. (2019). Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8(+) T Cells. Immunity 51, 285–297 e285. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

