Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins
- PMID: 35021064
- PMCID: PMC8809250
- DOI: 10.1016/j.cell.2021.12.021
Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins
Abstract
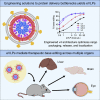
Methods to deliver gene editing agents in vivo as ribonucleoproteins could offer safety advantages over nucleic acid delivery approaches. We report the development and application of engineered DNA-free virus-like particles (eVLPs) that efficiently package and deliver base editor or Cas9 ribonucleoproteins. By engineering VLPs to overcome cargo packaging, release, and localization bottlenecks, we developed fourth-generation eVLPs that mediate efficient base editing in several primary mouse and human cell types. Using different glycoproteins in eVLPs alters their cellular tropism. Single injections of eVLPs into mice support therapeutic levels of base editing in multiple tissues, reducing serum Pcsk9 levels 78% following 63% liver editing, and partially restoring visual function in a mouse model of genetic blindness. In vitro and in vivo off-target editing from eVLPs was virtually undetected, an improvement over AAV or plasmid delivery. These results establish eVLPs as promising vehicles for therapeutic macromolecule delivery that combine key advantages of both viral and nonviral delivery.
Keywords: base editing; genome editing; in vivo delivery; ribonucleoproteins; therapeutic gene editing; virus-like particles.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare competing financial interests: S.B., A.R., and D.R.L. have filed patent applications on this work through the Broad Institute. K.M. is a consultant and equity holder of Verve Therapeutics and Variant Bio. K.P. is chief scientific officer of Polgenix, Inc. D.R.L. is a consultant and equity holder of Prime Medicine, Beam Therapeutics, Pairwise Plants, and Chroma Medicine, companies that use gene editing or genome engineering.
Figures


Comment in
-
Bringing gene therapy to where it's needed.Trends Mol Med. 2022 Mar;28(3):171-172. doi: 10.1016/j.molmed.2022.01.005. Epub 2022 Jan 24. Trends Mol Med. 2022. PMID: 35086771
-
Enhancing base editor delivery.Nat Rev Drug Discov. 2022 Mar;21(3):177. doi: 10.1038/d41573-022-00021-5. Nat Rev Drug Discov. 2022. PMID: 35102294 No abstract available.
-
Delivering therapeutic proteins in vivo by engineered DNA-free virus-like particles.Commun Biol. 2022 Apr 27;5(1):392. doi: 10.1038/s42003-022-03338-4. Commun Biol. 2022. PMID: 35477767 Free PMC article.
-
Engineered virus-like particles: paving the way for effective somatic genome editing.Signal Transduct Target Ther. 2022 Aug 12;7(1):279. doi: 10.1038/s41392-022-01089-6. Signal Transduct Target Ther. 2022. PMID: 35961953 Free PMC article. No abstract available.
References
-
- Abifadel M., Varret M., Rabès J.P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. - PubMed
-
- Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F30 EY033642/EY/NEI NIH HHS/United States
- P01 CA065493/CA/NCI NIH HHS/United States
- F30 EY029136/EY/NEI NIH HHS/United States
- RM1 HG009490/HG/NHGRI NIH HHS/United States
- R35 GM118062/GM/NIGMS NIH HHS/United States
- T32 EY024236/EY/NEI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
- R35 HL145203/HL/NHLBI NIH HHS/United States
- UG3 AI150551/AI/NIAID NIH HHS/United States
- T32 GM008620/GM/NIGMS NIH HHS/United States
- R01 EY009339/EY/NEI NIH HHS/United States
- T32 GM095450/GM/NIGMS NIH HHS/United States
- U01 AI142756/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

