Molecular dynamics simulations of the flexibility and inhibition of SARS-CoV-2 NSP 13 helicase
- PMID: 35021142
- PMCID: PMC8730789
- DOI: 10.1016/j.jmgm.2022.108122
Molecular dynamics simulations of the flexibility and inhibition of SARS-CoV-2 NSP 13 helicase
Abstract
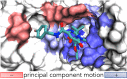
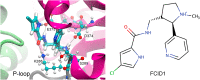
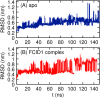
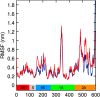
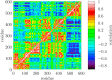
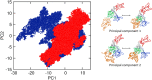
The helicase protein of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is both a good potential drug target and very flexible. The flexibility, and therefore its function, could be reduced through knowledge of these motions and identification of allosteric pockets. Using molecular dynamics simulations with enhanced sampling, we determined key modes of motion and sites on the protein that are at the interface between flexible domains of the proteins. We developed an approach to map the principal components of motion onto the surface of a potential binding pocket to help in the identification of allosteric sites.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
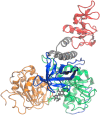




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

