A Collaborative Classroom Investigation of the Evolution of SABATH Methyltransferase Substrate Preference Shifts over 120 My of Flowering Plant History
- PMID: 35021222
- PMCID: PMC8890502
- DOI: 10.1093/molbev/msac007
A Collaborative Classroom Investigation of the Evolution of SABATH Methyltransferase Substrate Preference Shifts over 120 My of Flowering Plant History
Abstract
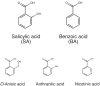
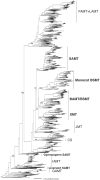
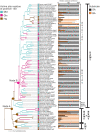
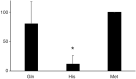
Next-generation sequencing has resulted in an explosion of available data, much of which remains unstudied in terms of biochemical function; yet, experimental characterization of these sequences has the potential to provide unprecedented insight into the evolution of enzyme activity. One way to make inroads into the experimental study of the voluminous data available is to engage students by integrating teaching and research in a college classroom such that eventually hundreds or thousands of enzymes may be characterized. In this study, we capitalize on this potential to focus on SABATH methyltransferase enzymes that have been shown to methylate the important plant hormone, salicylic acid (SA), to form methyl salicylate. We analyze data from 76 enzymes of flowering plant species in 23 orders and 41 families to investigate how widely conserved substrate preference is for SA methyltransferase orthologs. We find a high degree of conservation of substrate preference for SA over the structurally similar metabolite, benzoic acid, with recent switches that appear to be associated with gene duplication and at least three cases of functional compensation by paralogous enzymes. The presence of Met in active site position 150 is a useful predictor of SA methylation preference in SABATH methyltransferases but enzymes with other residues in the homologous position show the same substrate preference. Although our dense and systematic sampling of SABATH enzymes across angiosperms has revealed novel insights, this is merely the "tip of the iceberg" since thousands of sequences remain uncharacterized in this enzyme family alone.
Keywords: enzyme evolution; paralog compensation; plant methyltransferase evolution; substrate preference evolution.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Barkman TJ, Martins TR, Sutton E, Stout JT. 2007. Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol Biol Evol. 24(6):1320–1329. - PubMed
-
- Chaiprasongsuk M, Zhang C, Qian P, Chen XL, Li GL, Trigiano RN, Guo H, Chen F. 2018. Biochemical characterization in Norway spruce (Picea abies) of SABATH methyltransferases that methylate phytohormones. Phytochemistry 149:146–154. - PubMed
-
- Chen F, D’Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. 2003. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36(5):577–588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

