Functional coding haplotypes and machine-learning feature elimination identifies predictors of Methotrexate Response in Rheumatoid Arthritis patients
- PMID: 35022146
- PMCID: PMC8808170
- DOI: 10.1016/j.ebiom.2021.103800
Functional coding haplotypes and machine-learning feature elimination identifies predictors of Methotrexate Response in Rheumatoid Arthritis patients
Abstract
Background: Major challenges in large scale genetic association studies include not only the identification of causative single nucleotide polymorphisms (SNPs), but also accounting for SNP-SNP interactions. This study thus proposes a novel feature engineering approach integrating potentially functional coding haplotypes (pfcHap) with machine-learning (ML) feature selection to identify biologically meaningful, possibly causative genetic factors, that take into consideration potential SNP-SNP interactions within the pfcHap, to best predict for methotrexate (MTX) response in rheumatoid arthritis (RA) patients.
Methods: Exome sequencing from 349 RA patients were analysed, of which they were split into training and unseen test set. Inferred pfcHaps were combined with 30 non-genetic features to undergo ML recursive feature elimination with cross-validation using the training set. Predictive capacity and robustness of the selected features were assessed using six popular machine learning models through a train set cross-validation and evaluated in an unseen test set.
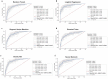
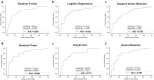
Findings: Significantly, 100 features (95 pfcHaps, 5 non-genetic factors) were identified to have good predictive performance (AUC: 0.776-0.828; Sensitivity: 0.656-0.813; Specificity: 0.684-0.868) across all six ML models in an unseen test dataset for the prediction of MTX response in RA patients.
Interpretation: Majority of the predictive pfcHap SNPs were predicted to be potentially functional and some of the genes in which the pfcHap resides in were identified to be associated with previously reported MTX/RA pathways.
Funding: Singapore Ministry of Health's National Medical Research Council (NMRC) [NMRC/CBRG/0095/2015; CG12Aug17; CGAug16M012; NMRC/CG/017/2013]; National Cancer Center Research Fund and block funding Duke-NUS Medical School.; Singapore Ministry of Education Academic Research Fund Tier 2 grant MOE2019-T2-1-138.
Keywords: Feature selection; Genetic polymorphism; Haplotypes; Machine learning; Methotrexate; Rheumatoid Arthritis.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest CGL, KPL, CCK, SSC, AJWL, and LJL declare that they have a pending Coversheet IP application.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

