Guide RNA acrobatics: the one-for-two shuffle
- PMID: 35022325
- PMCID: PMC8763051
- DOI: 10.1101/gad.349285.121
Guide RNA acrobatics: the one-for-two shuffle
Abstract
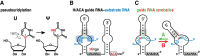
RNA modifications are crucial for the proper function of the RNAs. The sites of pseudouridines are often specified by dual hairpin guide RNAs, with one or both hairpins identifying a target uridine. In this issue of Genes & Development, Jády and colleagues (pp. 70-83) identify a novel mechanism by which a single guide RNA hairpin can specify two uridines adjacent to each other or separated by 1 nt; i.e., one for two or guide RNA acrobatics.
Keywords: RNA-guided RNA modification; RNA–RNA interaction; box H/ACA RNAs; guide RNA acrobatics; pseudouridine; pseudouridylation.
© 2022 Meier; Published by Cold Spring Harbor Laboratory Press.
Figures
Comment on
-
Guide RNA acrobatics: positioning consecutive uridines for pseudouridylation by H/ACA pseudouridylation loops with dual guide capacity.Genes Dev. 2022 Jan 1;36(1-2):70-83. doi: 10.1101/gad.349072.121. Epub 2021 Dec 16. Genes Dev. 2022. PMID: 34916304 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
