TWIST1 expression is associated with high-risk neuroblastoma and promotes primary and metastatic tumor growth
- PMID: 35022561
- PMCID: PMC8755726
- DOI: 10.1038/s42003-021-02958-6
TWIST1 expression is associated with high-risk neuroblastoma and promotes primary and metastatic tumor growth
Abstract
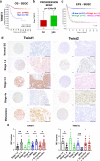
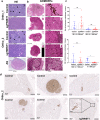
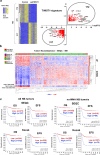
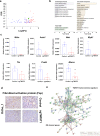
The embryonic transcription factors TWIST1/2 are frequently overexpressed in cancer, acting as multifunctional oncogenes. Here we investigate their role in neuroblastoma (NB), a heterogeneous childhood malignancy ranging from spontaneous regression to dismal outcomes despite multimodal therapy. We first reveal the association of TWIST1 expression with poor survival and metastasis in primary NB, while TWIST2 correlates with good prognosis. Secondly, suppression of TWIST1 by CRISPR/Cas9 results in a reduction of tumor growth and metastasis colonization in immunocompromised mice. Moreover, TWIST1 knockout tumors display a less aggressive cellular morphology and a reduced disruption of the extracellular matrix (ECM) reticulin network. Additionally, we identify a TWIST1-mediated transcriptional program associated with dismal outcome in NB and involved in the control of pathways mainly linked to the signaling, migration, adhesion, the organization of the ECM, and the tumor cells versus tumor stroma crosstalk. Taken together, our findings confirm TWIST1 as promising therapeutic target in NB.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

