Label2label: training a neural network to selectively restore cellular structures in fluorescence microscopy
- PMID: 35022745
- PMCID: PMC8918818
- DOI: 10.1242/jcs.258994
Label2label: training a neural network to selectively restore cellular structures in fluorescence microscopy
Abstract
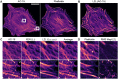
Immunofluorescence microscopy is routinely used to visualise the spatial distribution of proteins that dictates their cellular function. However, unspecific antibody binding often results in high cytosolic background signals, decreasing the image contrast of a target structure. Recently, convolutional neural networks (CNNs) were successfully employed for image restoration in immunofluorescence microscopy, but current methods cannot correct for those background signals. We report a new method that trains a CNN to reduce unspecific signals in immunofluorescence images; we name this method label2label (L2L). In L2L, a CNN is trained with image pairs of two non-identical labels that target the same cellular structure. We show that after L2L training a network predicts images with significantly increased contrast of a target structure, which is further improved after implementing a multiscale structural similarity loss function. Here, our results suggest that sample differences in the training data decrease hallucination effects that are observed with other methods. We further assess the performance of a cycle generative adversarial network, and show that a CNN can be trained to separate structures in superposed immunofluorescence images of two targets.
Keywords: Antibody labelling; Cellular structures; Content-aware image restoration; Convolutional neural networks; Fluorescence microscopy; Noise2noise.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Bradski, G. (2000). The OpenCV library. Dr. Dobb's J. Software Tools 120, 122-125.
-
- Caicedo, J. C., Roth, J., Goodman, A., Becker, T., Karhohs, K. W., Broisin, M., Molnar, C., Mcquin, C., Singh, S., Theis, F. J.et al. (2019). Evaluation of deep learning strategies for nucleus segmentation in fluorescence images. Cytometry Part A 95, 952-965. 10.1002/cyto.a.23863 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- EP/L016559/1/MRC_/Medical Research Council/United Kingdom
- 19-0238/AICR_/Worldwide Cancer Research/United Kingdom
- BB/T011602/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P02565X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K015583/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

