Functional Prediction of Biological Profile During Eutrophication in Marine Environment
- PMID: 35023908
- PMCID: PMC8744080
- DOI: 10.1177/11779322211063993
Functional Prediction of Biological Profile During Eutrophication in Marine Environment
Abstract
In the marine environment, coastal nutrient pollution and algal blooms are increasing in many coral reefs and surface waters around the world, leading to higher concentrations of dissolved organic carbon (DOC), nitrogen (N), phosphate (P), and sulfur (S) compounds. The adaptation of the marine microbiota to this stress involves evolutionary processes through mutations that can provide selective phenotypes. The aim of this in silico analysis is to elucidate the potential candidate hub proteins, biological processes, and key metabolic pathways involved in the pathogenicity of bacterioplankton during excess of nutrients. The analysis was carried out on the model organism Escherichia coli K-12, by adopting an analysis pipeline consisting of a set of packages from the Cystoscape platform. The results obtained show that the metabolism of carbon and sugars generally are the 2 driving mechanisms for the expression of virulence factors.
Keywords: Differentially expressed genes; copiotrophic species; functional analysis virulence; metabolic pathways.
© The Author(s) 2022.
Conflict of interest statement
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures


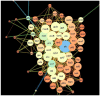
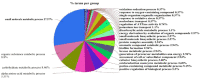

Similar articles
-
Excess labile carbon promotes the expression of virulence factors in coral reef bacterioplankton.ISME J. 2018 Jan;12(1):59-76. doi: 10.1038/ismej.2017.142. Epub 2017 Sep 12. ISME J. 2018. PMID: 28895945 Free PMC article.
-
Metabolic Roles of Uncultivated Bacterioplankton Lineages in the Northern Gulf of Mexico "Dead Zone".mBio. 2017 Sep 12;8(5):e01017-17. doi: 10.1128/mBio.01017-17. mBio. 2017. PMID: 28900024 Free PMC article.
-
Sensitivity of bacterioplankton nitrogen metabolism to eutrophication in sub-tropical coastal waters of Key West, Florida.Mar Pollut Bull. 2008 May;56(5):913-26. doi: 10.1016/j.marpolbul.2008.01.030. Epub 2008 Mar 10. Mar Pollut Bull. 2008. PMID: 18331746
-
Flow injection analysis as a tool for enhancing oceanographic nutrient measurements--a review.Anal Chim Acta. 2013 Nov 25;803:15-40. doi: 10.1016/j.aca.2013.06.015. Epub 2013 Jun 20. Anal Chim Acta. 2013. PMID: 24216194 Review.
-
Harmful algal blooms: causes, impacts and detection.J Ind Microbiol Biotechnol. 2003 Jul;30(7):383-406. doi: 10.1007/s10295-003-0074-9. Epub 2003 Jul 30. J Ind Microbiol Biotechnol. 2003. PMID: 12898390 Review.
Cited by
-
Effect of Climate on Bacterial and Archaeal Diversity of Moroccan Marine Microbiota.Microorganisms. 2022 Aug 10;10(8):1622. doi: 10.3390/microorganisms10081622. Microorganisms. 2022. PMID: 36014042 Free PMC article.
References
-
- Huang Z, Liu F, Luo P, et al.. Pilot-scale constructed wetlands for swine wastewater treatment: microbial community analysis in bacterioplankton and epiphyton and options for resource recovery. J Water Process Eng 2020;37:101466. doi:10.1016/j.jwpe.2020.101466. - DOI
LinkOut - more resources
Full Text Sources

