Mechanism of action of hepatitis B virus S antigen transport-inhibiting oligonucleotide polymer, STOPS, molecules
- PMID: 35024245
- PMCID: PMC8717253
- DOI: 10.1016/j.omtn.2021.12.013
Mechanism of action of hepatitis B virus S antigen transport-inhibiting oligonucleotide polymer, STOPS, molecules
Abstract
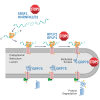
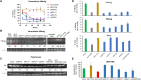
A functional cure of chronic hepatitis B requires eliminating the hepatitis B virus (HBV)-encoded surface antigen (HBsAg), which can suppress immune responses. STOPS are phosphorothioated single-stranded oligonucleotides containing novel chemistries that significantly reduce HBsAgs produced by HBV-infected liver cells. The STOPS molecule ALG-10000 functions inside cells to reduce the levels of multiple HBV-encoded molecules. However, it does not bind HBV molecules. An affinity resin coupled with ALG-10000 was found to bind several proteins from liver cells harboring replicating HBV. Silencing RNAs targeting host factors SRSF1, HNRNPA2B1, GRP78 (HspA5), RPLP1, and RPLP2 reduced HBsAg levels and other HBV molecules that are concomitantly reduced by STOPS. Host proteins RPLP1/RPLP2 and GRP78 function in the translation of membrane proteins, protein folding, and degradation. ALG-10000 and the knockdowns of RPLP1/2 and GRP78 decreased the levels of HBsAg and increased their ubiquitination and proteasome degradation. GRP78, RPLP1, and RPLP2 affected HBsAg production only when HBsAg was expressed with HBV regulatory sequences, suggesting that HBV has evolved to engage with these STOPS-interacting molecules. The STOPS inhibition of HBsAg levels in HBV-infected cells occurs by sequestering cellular proteins needed for proper expression and folding of HBsAg.
Keywords: GRP78; HBV S antigen; RPLP1; RPLP2; antiviral; chronic hepatitis B; functional cure; hepatitis B virus; host factors; nucleic acid polymers.
© 2021 The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures







References
-
- World Health Organization . World Health Organization; 2021. Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections. ISBN: 978-92-4-002707-7.
-
- Tang L.S.Y., Covert E., Wilson E., Kottilil S. Chronic hepatitis B infection, a review. JAMA 319:1802-1813.6. Yip TC, Lok AS. 2020. How do we determine whether a functional cure of HBV infection has been achieved? Clin. Gastroenterol. Hepatol. 2018;18:548–550.
LinkOut - more resources
Full Text Sources
Miscellaneous

