Supramolecular assembly of protein building blocks: from folding to function
- PMID: 35024976
- PMCID: PMC8755899
- DOI: 10.1186/s40580-021-00294-3
Supramolecular assembly of protein building blocks: from folding to function
Abstract
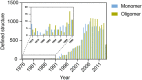
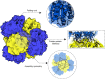
Several phenomena occurring throughout the life of living things start and end with proteins. Various proteins form one complex structure to control detailed reactions. In contrast, one protein forms various structures and implements other biological phenomena depending on the situation. The basic principle that forms these hierarchical structures is protein self-assembly. A single building block is sufficient to create homogeneous structures with complex shapes, such as rings, filaments, or containers. These assemblies are widely used in biology as they enable multivalent binding, ultra-sensitive regulation, and compartmentalization. Moreover, with advances in the computational design of protein folding and protein-protein interfaces, considerable progress has recently been made in the de novo design of protein assemblies. Our review presents a description of the components of supramolecular protein assembly and their application in understanding biological phenomena to therapeutics.
Keywords: Protein design; Protein folding; Protein–protein interaction; Supramolecular assembly.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
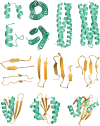



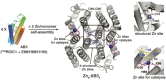

References
-
- Pieters BJ, et al. Natural supramolecular protein assemblies. Chem. Soc. Rev. 2016;45(1):24–39. - PubMed
-
- Garcia-Seisdedos H, et al. Proteins evolve on the edge of supramolecular self-assembly. Nature. 2017;548(7666):244–247. - PubMed
-
- Marsh JA, Teichmann SA. Structure, Dynamics, Assembly, and Evolution of Protein Complexes. Annu. Rev. Biochem. 2015;84(84):551–575. - PubMed
-
- DeGrado WF, Regan L, Ho SP. The design of a four-helix bundle protein. Cold Spring Harb. Symp. Quant. Biol. 1987;52:521–526. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
