RECON syndrome is a genome instability disorder caused by mutations in the DNA helicase RECQL1
- PMID: 35025765
- PMCID: PMC8884905
- DOI: 10.1172/JCI147301
RECON syndrome is a genome instability disorder caused by mutations in the DNA helicase RECQL1
Abstract
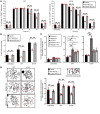
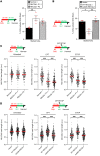
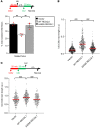
Despite being the first homolog of the bacterial RecQ helicase to be identified in humans, the function of RECQL1 remains poorly characterized. Furthermore, unlike other members of the human RECQ family of helicases, mutations in RECQL1 have not been associated with a genetic disease. Here, we identify 2 families with a genome instability disorder that we have named RECON (RECql ONe) syndrome, caused by biallelic mutations in the RECQL gene. The affected individuals had short stature, progeroid facial features, a hypoplastic nose, xeroderma, and skin photosensitivity and were homozygous for the same missense mutation in RECQL1 (p.Ala459Ser), located within its zinc binding domain. Biochemical analysis of the mutant RECQL1 protein revealed that the p.A459S missense mutation compromised its ATPase, helicase, and fork restoration activity, while its capacity to promote single-strand DNA annealing was largely unaffected. At the cellular level, this mutation in RECQL1 gave rise to a defect in the ability to repair DNA damage induced by exposure to topoisomerase poisons and a failure of DNA replication to progress efficiently in the presence of abortive topoisomerase lesions. Taken together, RECQL1 is the fourth member of the RecQ family of helicases to be associated with a human genome instability disorder.
Keywords: Cell Biology; DNA repair; Genetic diseases; Genetic instability; Genetics.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

