Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19
- PMID: 35026155
- PMCID: PMC8712267
- DOI: 10.1016/j.celrep.2021.110271
Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19
Abstract
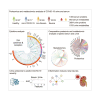
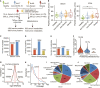
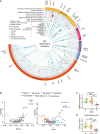
The utility of the urinary proteome in infectious diseases remains unclear. Here, we analyzed the proteome and metabolome of urine and serum samples from patients with COVID-19 and healthy controls. Our data show that urinary proteins effectively classify COVID-19 by severity. We detect 197 cytokines and their receptors in urine, but only 124 in serum using TMT-based proteomics. The decrease in urinary ESCRT complex proteins correlates with active SARS-CoV-2 replication. The downregulation of urinary CXCL14 in severe COVID-19 cases positively correlates with blood lymphocyte counts. Integrative multiomics analysis suggests that innate immune activation and inflammation triggered renal injuries in patients with COVID-19. COVID-19-associated modulation of the urinary proteome offers unique insights into the pathogenesis of this disease. This study demonstrates the added value of including the urinary proteome in a suite of multiomics analytes in evaluating the immune pathobiology and clinical course of COVID-19 and, potentially, other infectious diseases.
Keywords: COVID-19; CXCL14; ESCRT super-complex; metabolomics; proteomics; renal injury; serum; urine.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interest The research group of T.G. is partly supported by Pressure Biosciences. T.G. and Y. Zhu are shareholders of Westlake Omics. W.L., X.Y., N.X., W.G., and X. Zhan are currently employees of Westlake Omics. S.Q., C.Z., and H.L. are employees of Calibra Lab at DIAN Diagnostics. The remaining authors declare no competing interests.
Figures



References
-
- Biedroń R., Ciszek M., Tokarczyk M., Bobek M., Kurnyta M., Słominska E.M., Smoleński R.T., Marcinkiewicz J. 1-Methylnicotinamide and nicotinamide: two related anti-inflammatory agents that differentially affect the functions of activated macrophages. Arch. Immunol. Ther. Exp. (Warsz) 2008;56:127–134. doi: 10.1007/s00005-008-0009-2. - DOI - PMC - PubMed
-
- Block F., Schwarz M. Expression of GFAP in the striatum and its projection areas in response to striatal quinolinic acid lesion in rats. Neuroreport. 1994;5:2237–2240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

