The genetic architecture of pediatric cardiomyopathy
- PMID: 35026164
- PMCID: PMC8874151
- DOI: 10.1016/j.ajhg.2021.12.006
The genetic architecture of pediatric cardiomyopathy
Abstract
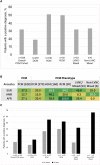
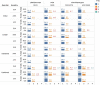
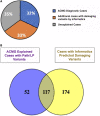
To understand the genetic contribution to primary pediatric cardiomyopathy, we performed exome sequencing in a large cohort of 528 children with cardiomyopathy. Using clinical interpretation guidelines and targeting genes implicated in cardiomyopathy, we identified a genetic cause in 32% of affected individuals. Cardiomyopathy sub-phenotypes differed by ancestry, age at diagnosis, and family history. Infants < 1 year were less likely to have a molecular diagnosis (p < 0.001). Using a discovery set of 1,703 candidate genes and informatic tools, we identified rare and damaging variants in 56% of affected individuals. We see an excess burden of damaging variants in affected individuals as compared to two independent control sets, 1000 Genomes Project (p < 0.001) and SPARK parental controls (p < 1 × 10-16). Cardiomyopathy variant burden remained enriched when stratified by ancestry, variant type, and sub-phenotype, emphasizing the importance of understanding the contribution of these factors to genetic architecture. Enrichment in this discovery candidate gene set suggests multigenic mechanisms underlie sub-phenotype-specific causes and presentations of cardiomyopathy. These results identify important information about the genetic architecture of pediatric cardiomyopathy and support recommendations for clinical genetic testing in children while illustrating differences in genetic architecture by age, ancestry, and sub-phenotype and providing rationale for larger studies to investigate multigenic contributions.
Keywords: ancestry; bioinformatics; clinical interpretation; exome; heart; infant; molecular diagnosis; variant.
Copyright © 2021 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.W.R. is a consultant for Amgen, Bayer, Novartis, and Abiomed. W.K.C. is on the scientific advisory board for the Regeneron Genetics Center. S.E.L. is a consultant for Tenaya Therapeutics and Bayer and on an advisory board for Myokardia.
Figures




References
-
- Lipshultz S.E., Orav E.J., Wilkinson J.D., Towbin J.A., Messere J.E., Lowe A.M., Sleeper L.A., Cox G.F., Hsu D.T., Canter C.E., et al. Risk stratification at diagnosis for children with hypertrophic cardiomyopathy: an analysis of data from the Pediatric Cardiomyopathy Registry. Lancet. 2013;382:1889–1897. - PMC - PubMed
-
- Pahl E., Sleeper L.A., Canter C.E., Hsu D.T., Lu M., Webber S.A., Colan S.D., Kantor P.F., Everitt M.D., Towbin J.A., et al. Incidence of and risk factors for sudden cardiac death in children with dilated cardiomyopathy: a report from the Pediatric Cardiomyopathy Registry. J. Am. Coll. Cardiol. 2012;59:607–615. - PMC - PubMed
-
- Webber S.A., Lipshultz S.E., Sleeper L.A., Lu M., Wilkinson J.D., Addonizio L.J., Canter C.E., Colan S.D., Everitt M.D., Jefferies J.L., et al. Outcomes of restrictive cardiomyopathy in childhood and the influence of phenotype: a report from the Pediatric Cardiomyopathy Registry. Circulation. 2012;126:1237–1244. - PubMed
-
- Daubeney P.E., Nugent A.W., Chondros P., Carlin J.B., Colan S.D., Cheung M., Davis A.M., Chow C.W., Weintraub R.G., National Australian Childhood Cardiomyopathy Study Clinical features and outcomes of childhood dilated cardiomyopathy: results from a national population-based study. Circulation. 2006;114:2671–2678. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

