Predicting sepsis severity at first clinical presentation: The role of endotypes and mechanistic signatures
- PMID: 35027333
- PMCID: PMC8808161
- DOI: 10.1016/j.ebiom.2021.103776
Predicting sepsis severity at first clinical presentation: The role of endotypes and mechanistic signatures
Abstract
Background: Inter-individual variability during sepsis limits appropriate triage of patients. Identifying, at first clinical presentation, gene expression signatures that predict subsequent severity will allow clinicians to identify the most at-risk groups of patients and enable appropriate antibiotic use.
Methods: Blood RNA-Seq and clinical data were collected from 348 patients in four emergency rooms (ER) and one intensive-care-unit (ICU), and 44 healthy controls. Gene expression profiles were analyzed using machine learning and data mining to identify clinically relevant gene signatures reflecting disease severity, organ dysfunction, mortality, and specific endotypes/mechanisms.
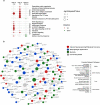
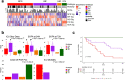
Findings: Gene expression signatures were obtained that predicted severity/organ dysfunction and mortality in both ER and ICU patients with accuracy/AUC of 77-80%. Network analysis revealed these signatures formed a coherent biological program, with specific but overlapping mechanisms/pathways. Given the heterogeneity of sepsis, we asked if patients could be assorted into discrete groups with distinct mechanisms (endotypes) and varying severity. Patients with early sepsis could be stratified into five distinct and novel mechanistic endotypes, named Neutrophilic-Suppressive/NPS, Inflammatory/INF, Innate-Host-Defense/IHD, Interferon/IFN, and Adaptive/ADA, each based on ∼200 unique gene expression differences, and distinct pathways/mechanisms (e.g., IL6/STAT3 in NPS). Endotypes had varying overall severity with two severe (NPS/INF) and one relatively benign (ADA) groupings, consistent with reanalysis of previous endotype studies. A 40 gene-classification tool (accuracy=96%) and several gene-pairs (accuracy=89-97%) accurately predicted endotype status in both ER and ICU validation cohorts.
Interpretation: The severity and endotype signatures indicate that distinct immune signatures precede the onset of severe sepsis and lethality, providing a method to triage early sepsis patients.
Keywords: Cellular reprogramming; Endotypes; Gene signatures & biomarkers; Sepsis; Severe sepsis; Translational medicine.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests RH has filed the CR and endotype signatures for patent protection and licenced this to Sepset Biotherapeutics Inc., a Vancouver company in which he has a significant ownership position. AB and GCF have filed the endotype signatures for patent protection and licenced this to Sepset Biotherapeutics Inc. Other authors have nothing to disclose.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

