53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polα-primase fill-in synthesis
- PMID: 35027730
- PMCID: PMC8849574
- DOI: 10.1038/s41556-021-00812-9
53BP1-shieldin-dependent DSB processing in BRCA1-deficient cells requires CST-Polα-primase fill-in synthesis
Abstract
The efficacy of poly(ADP)-ribose polymerase 1 inhibition (PARPi) in BRCA1-deficient cells depends on 53BP1 and shieldin, which have been proposed to limit single-stranded DNA at double-strand breaks (DSBs) by blocking resection and/or through CST-Polα-primase-mediated fill-in. We show that primase (like 53BP1-shieldin and CST-Polα) promotes radial chromosome formation in PARPi-treated BRCA1-deficient cells and demonstrate shieldin-CST-Polα-primase-dependent incorporation of BrdU at DSBs. In the absence of 53BP1 or shieldin, radial formation in BRCA1-deficient cells was restored by the tethering of CST near DSBs, arguing that in this context, shieldin acts primarily by recruiting CST. Furthermore, a SHLD1 mutant defective in CST binding (SHLD1Δ) was non-functional in BRCA1-deficient cells and its function was restored after reconnecting SHLD1Δ to CST. Interestingly, at dysfunctional telomeres and at DNA breaks in class switch recombination where CST has been implicated, SHLD1Δ was fully functional, perhaps because these DNA ends carry CST recognition sites that afford SHLD1-independent binding of CST. These data establish that in BRCA1-deficient cells, CST-Polα-primase is the major effector of shieldin-dependent DSB processing.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


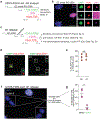

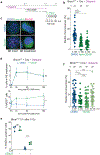




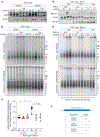



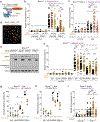


References
-
- Hustedt N & Durocher D The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9 (2016). - PubMed
-
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR & Bishop DK The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 275, 23899–23903 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

