HLA-independent T cell receptors for targeting tumors with low antigen density
- PMID: 35027758
- PMCID: PMC9469647
- DOI: 10.1038/s41591-021-01621-1
HLA-independent T cell receptors for targeting tumors with low antigen density
Abstract
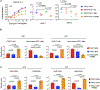
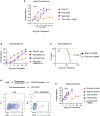
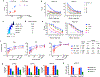
Chimeric antigen receptors (CARs) are receptors for antigen that direct potent immune responses. Tumor escape associated with low target antigen expression is emerging as one potential limitation of their efficacy. Here we edit the TRAC locus in human peripheral blood T cells to engage cell-surface targets through their T cell receptor-CD3 complex reconfigured to utilize the same immunoglobulin heavy and light chains as a matched CAR. We demonstrate that these HLA-independent T cell receptors (HIT receptors) consistently afford high antigen sensitivity and mediate tumor recognition beyond what CD28-based CARs, the most sensitive design to date, can provide. We demonstrate that the functional persistence of HIT T cells can be augmented by constitutive coexpression of CD80 and 4-1BBL. Finally, we validate the increased antigen sensitivity afforded by HIT receptors in xenograft mouse models of B cell leukemia and acute myeloid leukemia, targeting CD19 and CD70, respectively. Overall, HIT receptors are well suited for targeting cell surface antigens of low abundance.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures
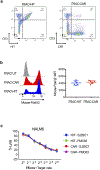


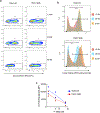
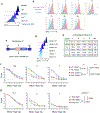

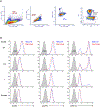




References
Methods-Only References
-
- Navarrete-Perea J, Yu Q, Gygi SP & Paulo JA Streamlined Tandem Mass Tag (SL-TMT) Protocol: An Efficient Strategy for Quantitative (Phospho)proteome Profiling Using Tandem Mass Tag-Synchronous Precursor Selection-MS3. Journal of proteome research 17, 2226–2236, doi:10.1021/acs.jproteome.8b00217 (2018). - DOI - PMC - PubMed
-
- Zucchetti AE, Bataille L, Carpier JM, Dogniaux S, San Roman-Jouve M, Maurin M, Stuck MW, Rios RM, Baldari CT, Pazour GJ, Hivroz C Tethering of vesicles to the Golgi by GMAP210 controls LAT delivery to the immune synapse. Nat Commun. 10, 2864. doi:10.1038/s41467-019-10891-w (2019). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

