Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO
- PMID: 35027765
- PMCID: PMC8828471
- DOI: 10.1038/s41592-021-01343-9
Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO
Abstract
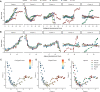
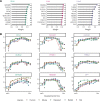
Factor analysis is a widely used method for dimensionality reduction in genome biology, with applications from personalized health to single-cell biology. Existing factor analysis models assume independence of the observed samples, an assumption that fails in spatio-temporal profiling studies. Here we present MEFISTO, a flexible and versatile toolbox for modeling high-dimensional data when spatial or temporal dependencies between the samples are known. MEFISTO maintains the established benefits of factor analysis for multimodal data, but enables the performance of spatio-temporally informed dimensionality reduction, interpolation, and separation of smooth from non-smooth patterns of variation. Moreover, MEFISTO can integrate multiple related datasets by simultaneously identifying and aligning the underlying patterns of variation in a data-driven manner. To illustrate MEFISTO, we apply the model to different datasets with spatial or temporal resolution, including an evolutionary atlas of organ development, a longitudinal microbiome study, a single-cell multi-omics atlas of mouse gastrulation and spatially resolved transcriptomics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

